2 xtitle: coherence & propositions
observations in :schizophrenia: threads
date: 2025-07-27 22:18:07
eval output M11, normalised to obs, distance ceiling = outliers removed.
2.1 method (M11)
To compute distances we queried a corpus for matching conditions where certain (assumed) determiners appear before similar nouns. In M11 (removed…)
This distance should give us information structural evidence of how strong these noun occurences are connected, i.e. if a noun appears out of the blue mostly or if it somewhere before has been introduced to the audience. In information structure definitions this would be termed with given and new information (Prince 1981).
2.2 legende
| variable | explanation | values |
|---|---|---|
| target | corpus | obs,ref |
| q | condition | a,b,c,d,e,f |
| det | antecedent POS==DET | TRUE,FALSE |
| q:a | query condition | .* |
| q:b | query condition | this,that,those,these |
| q:c | query condition | the |
| q:d | query condition | a,an,any,some |
| q:e | query condition | my |
| q:f | query condition | his,her,their,your |
2.3 anova analysis
2.3.1 anova plain, formula: [dist_rel_obs ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 3.6884e+09 3688441713 86368.2495 < 2.2e-16 ***
## q 5 4.5563e+08 91126214 2133.8040 < 2.2e-16 ***
## det 1 2.8130e+07 28129559 658.6794 < 2.2e-16 ***
## target:q 5 2.6402e+06 528048 12.3647 5.105e-12 ***
## target:det 1 2.3260e+06 2325954 54.4643 1.584e-13 ***
## q:det 1 2.7700e+06 2769968 64.8613 8.044e-16 ***
## target:q:det 1 4.3320e+03 4332 0.1014 0.7501
## Residuals 1045139 4.4634e+10 42706
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 12.3.2 anova of linear regression model: [anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 6094426 6094426 1 718328 185.7599 < 2.2e-16 ***
## q 42974986 8594997 5 1037780 261.9780 < 2.2e-16 ***
## det 372221 372221 1 1037864 11.3454 0.0007564 ***
## target:q 1766151 353230 5 1037779 10.7666 2.27e-10 ***
## target:det 92612 92612 1 1037867 2.8228 0.0929316 .
## q:det 326995 326995 1 1037796 9.9669 0.0015938 **
## target:q:det 137650 137650 1 1037795 4.1956 0.0405288 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 12.3.3 linear regression coefficients, formula: [dist_rel_obs ~ target*q*det+(1|lemma)+(1|aut_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method ['lmerModLmerTest']
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 13864438
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -6.3215 -0.4316 -0.0874 0.1283 12.4351
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 40316 200.8
## lemma (Intercept) 42511 206.2
## Residual 32808 181.1
## Number of obs: 1045155, groups: aut_id, 4533; lemma, 3652
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.870e+02 4.900e+00 6.838e+03 58.565 < 2e-16 ***
## targetref -8.842e+01 3.051e+00 7.021e+04 -28.978 < 2e-16 ***
## qb 3.566e+01 7.380e+00 1.036e+06 4.833 1.35e-06 ***
## qc 6.937e+01 1.268e+00 1.038e+06 54.728 < 2e-16 ***
## qd 5.704e+01 1.325e+00 1.038e+06 43.053 < 2e-16 ***
## qe 7.522e+01 1.447e+00 1.038e+06 51.975 < 2e-16 ***
## qf 6.010e+01 1.805e+00 1.037e+06 33.291 < 2e-16 ***
## detTRUE 3.154e+00 6.295e-01 1.042e+06 5.011 5.42e-07 ***
## targetref:qb 2.973e+01 1.105e+01 1.038e+06 2.690 0.007140 **
## targetref:qc 1.748e+01 6.460e+00 1.038e+06 2.706 0.006813 **
## targetref:qd 3.302e+01 6.156e+00 1.037e+06 5.363 8.18e-08 ***
## targetref:qe -2.499e+01 8.090e+00 1.038e+06 -3.090 0.002005 **
## targetref:qf -1.280e+02 3.760e+01 1.038e+06 -3.404 0.000663 ***
## targetref:detTRUE -3.951e+00 1.561e+00 1.043e+06 -2.531 0.011370 *
## qb:detTRUE 1.201e+01 7.839e+00 1.036e+06 1.532 0.125614
## targetref:qb:detTRUE 4.436e+01 2.166e+01 1.038e+06 2.048 0.040529 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 8 columns / coefficients2.4 plots
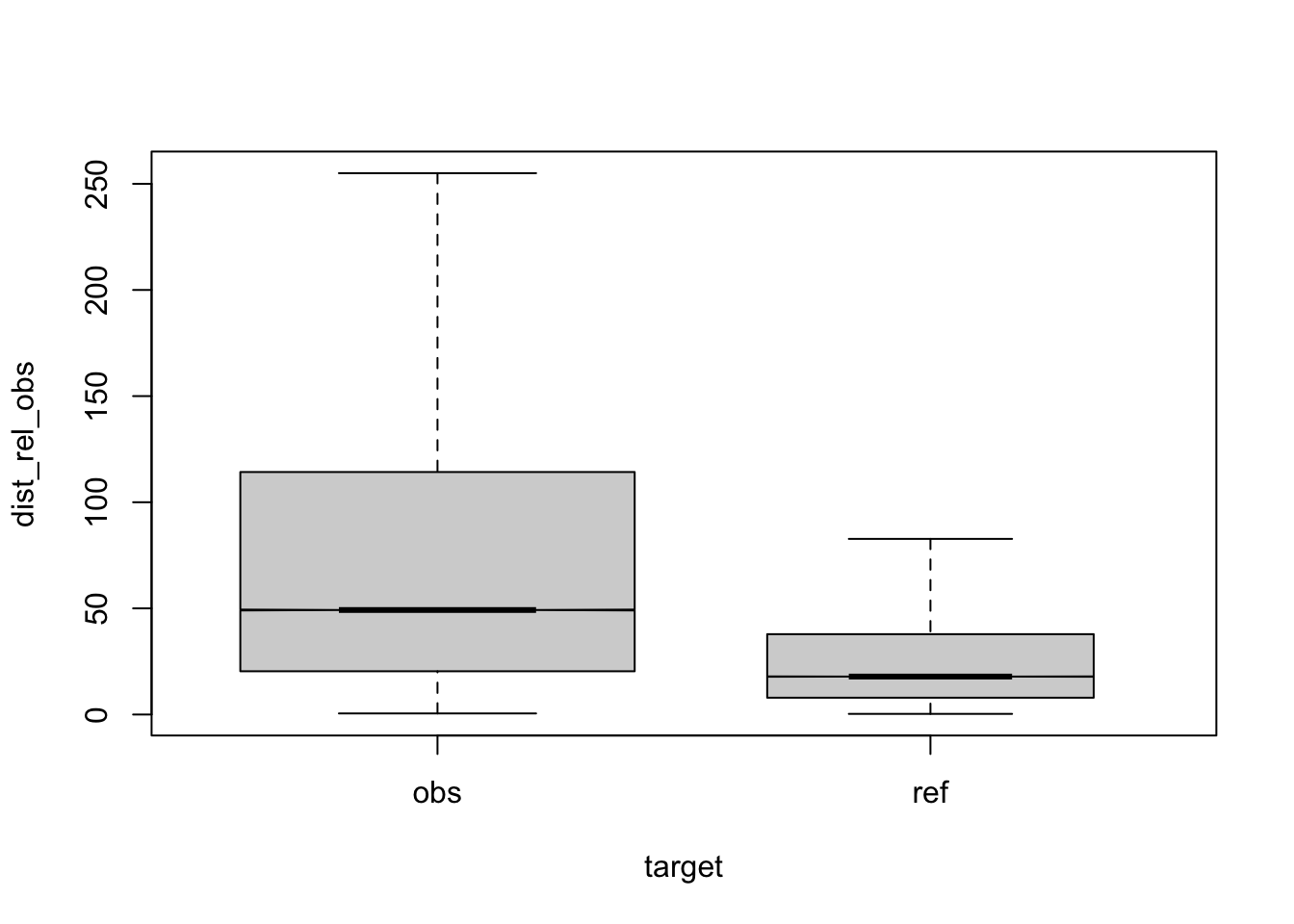
Figure 2.1: compare distances by corpus, normalised to obs, distance ceiling = outliers removed
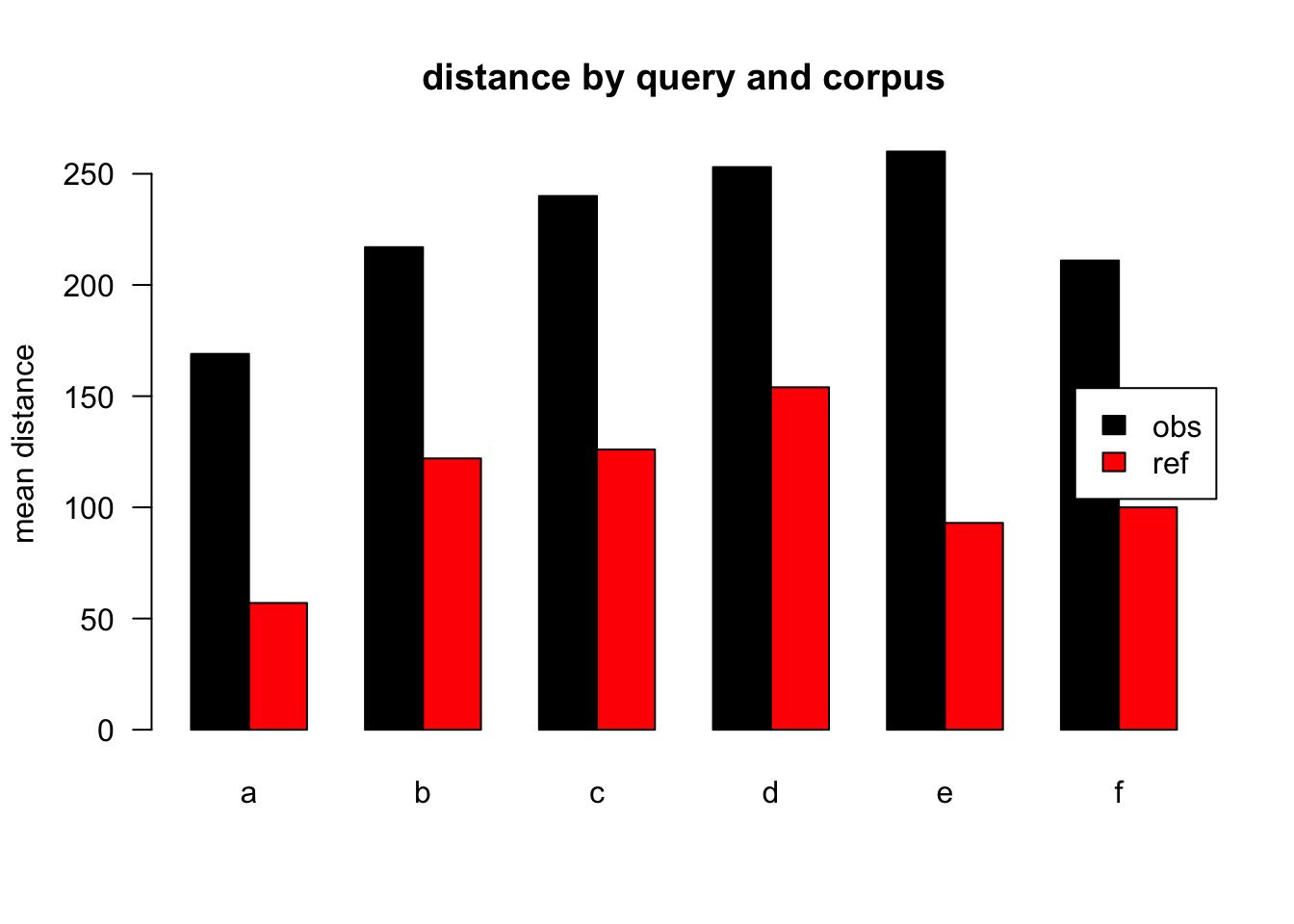
Figure 2.2: mean distances over query/corpus, normalised to obs, distance ceiling = outliers removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 557479 | 169 | 77 |
| ref | a | 394838 | 57 | 25 |
| obs | b | 5904 | 217 | 120 |
| ref | b | 747 | 122 | 44 |
| obs | c | 26883 | 240 | 138 |
| ref | c | 1033 | 126 | 73 |
| obs | d | 25400 | 253 | 149 |
| ref | d | 1041 | 154 | 106 |
| obs | e | 19228 | 260 | 149 |
| ref | e | 656 | 93 | 79 |
| obs | f | 11920 | 211 | 111 |
| ref | f | 26 | 100 | 126 |
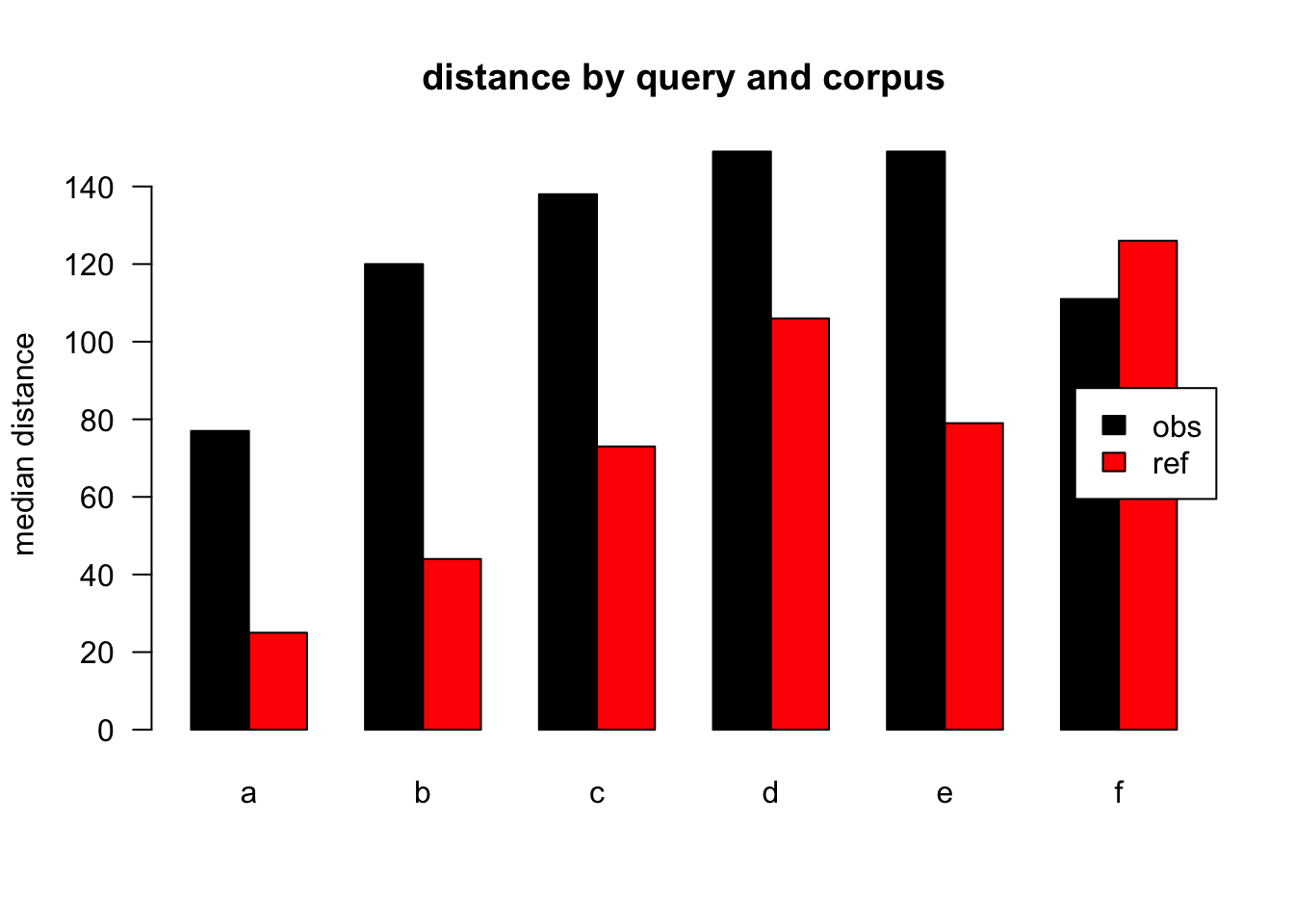
Figure 2.3: median distances over query/corpus, normalised to obs, distance ceiling = outliers removed
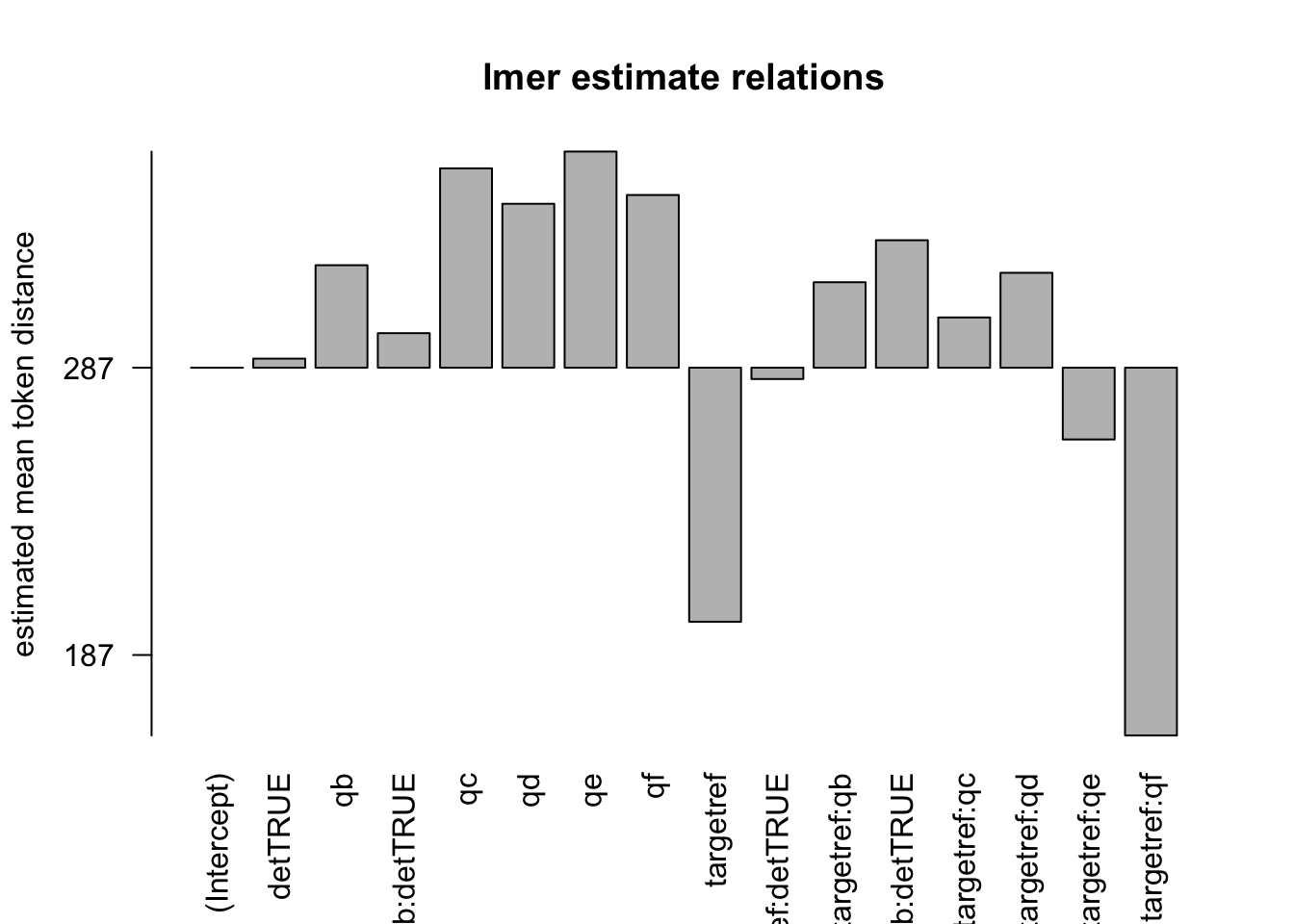
Figure 2.4: distances relation, normalised to obs, distance ceiling = outliers removed
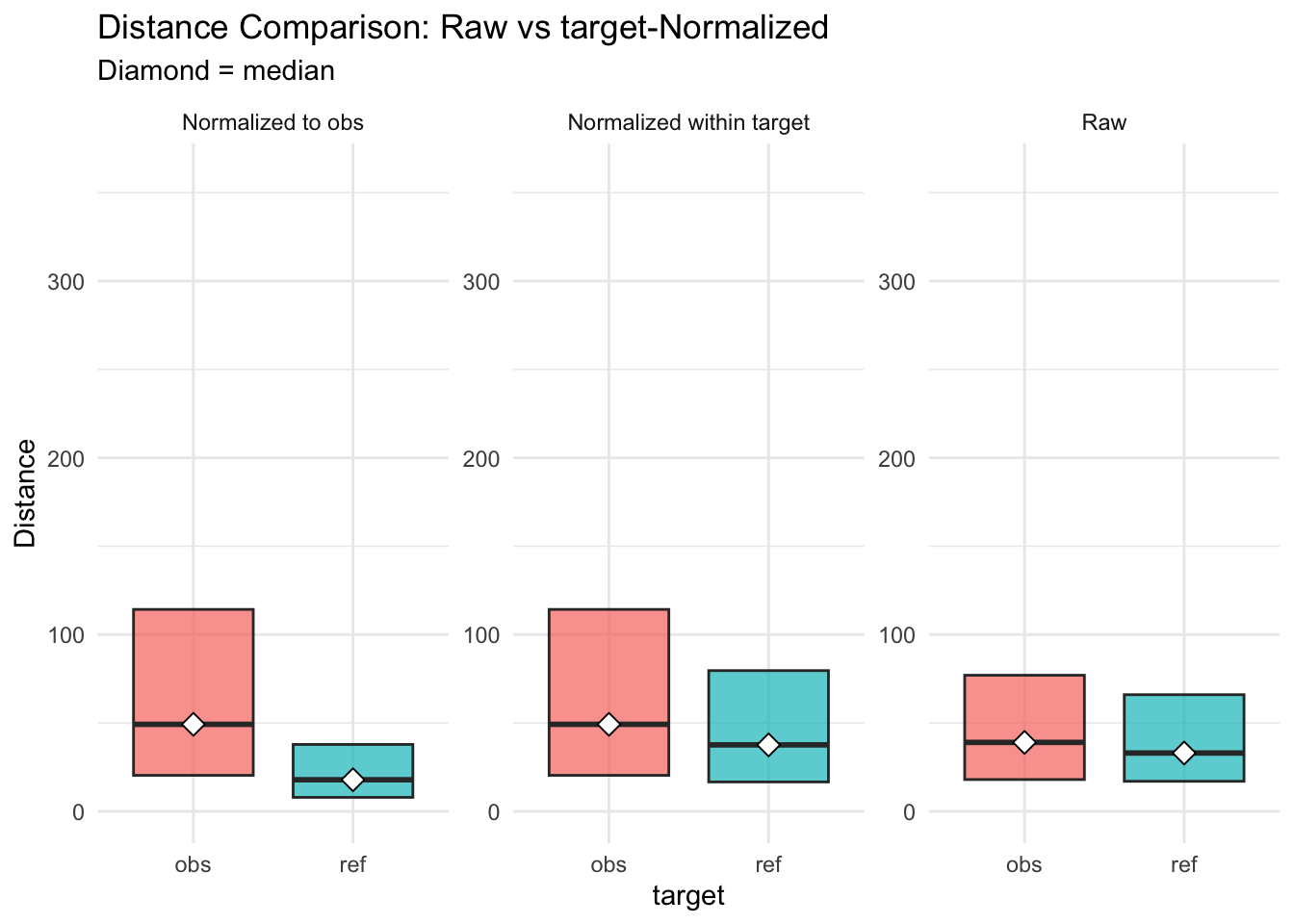
Figure 2.5: distances normalised vs. raw