3 appendix
overall wordcount of paper: 3071.
3.1 legende
| variable | explanation | values |
|---|---|---|
| target | corpus | obs,ref |
| q | condition | a,b,c,d,e,f |
| det | antecedent POS==DET | TRUE,FALSE |
| aut_id | author | author hash |
| lemma | lemma | noun lemma |
| range | url range of distance devised | 1..maxlength(urlthread) |
| embed.score | semantic similarity score lemma vs. thread | 0..1 |
| q:a | query condition | .* |
| q:b | query condition | this,that,those,these |
| q:c | query condition | the |
| q:d | query condition | a,an,any,some |
| q:e | query condition | my |
| q:f | query condition | his,her,their,your |
3.2 evaluation model: 1
3.2.2 parameter setting
## value
## norm_target _rel_obs
## det.t TRUE
## limit TRUE
## author TRUE
## url TRUE
## embed1 TRUE
## embed2 f
## range1 TRUE
## range2 f
## rel TRUE
## lme FALSE
## lemma FALSE3.2.3 anova analysis
3.2.3.1 anova plain
formula: [dist_rel_obs ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 452303747 452303747 7336.4625 < 2.2e-16 ***
## q 5 12320667 2464133 39.9688 < 2.2e-16 ***
## det 1 1636109 1636109 26.5380 2.588e-07 ***
## target:q 5 2747371 549474 8.9126 1.786e-08 ***
## target:det 1 251297 251297 4.0761 0.043496 *
## q:det 2 905292 452646 7.3420 0.000648 ***
## target:q:det 1 717222 717222 11.6335 0.000648 ***
## Residuals 126209 7780971239 61651
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.2.3.2 anova of linear regression model
[anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 1144219 1144219 1 3519 23.4567 1.333e-06 ***
## q 737483 147497 5 122421 3.0237 0.0098706 **
## det 12165 12165 1 118425 0.2494 0.6175055
## range 50399647 50399647 1 1025 1033.2042 < 2.2e-16 ***
## embed.score 25101881 25101881 1 122690 514.5942 < 2.2e-16 ***
## target:q 776335 155267 5 123486 3.1830 0.0070933 **
## target:det 541078 541078 1 123325 11.0922 0.0008672 ***
## q:det 359520 179760 2 120804 3.6851 0.0250971 *
## target:q:det 219844 219844 1 123315 4.5068 0.0337615 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.2.3.3 linear regression coefficients
formula: [dist_rel_obs ~ target*q*det+(1|aut_id)+range+(embed.score)+(1|url_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 1727639
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.8643 -0.5282 -0.1721 0.2469 6.9244
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 2856 53.44
## url_id (Intercept) 8187 90.48
## Residual 48780 220.86
## Number of obs: 126226, groups: aut_id, 8238; url_id, 2145
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 4.625e+02 5.159e+00 8.969e+03 89.651 < 2e-16 ***
## targetref -4.342e+01 6.299e+00 1.300e+03 -6.893 8.50e-12 ***
## qb -2.013e+01 1.527e+01 1.218e+05 -1.318 0.187483
## qc -2.207e+01 5.499e+00 1.226e+05 -4.014 5.98e-05 ***
## qd -3.178e+01 2.225e+02 1.184e+05 -0.143 0.886426
## qe 2.492e+01 3.835e+00 1.247e+05 6.498 8.14e-11 ***
## qf -1.891e+01 4.892e+00 1.244e+05 -3.866 0.000111 ***
## det 1.273e+01 4.774e+00 1.229e+05 2.667 0.007662 **
## range -5.810e-02 1.808e-03 1.025e+03 -32.143 < 2e-16 ***
## embed.score -1.829e+02 8.061e+00 1.227e+05 -22.685 < 2e-16 ***
## targetref:qb 1.862e+01 1.719e+01 1.225e+05 1.083 0.278599
## targetref:qc 2.281e+01 1.279e+01 1.237e+05 1.784 0.074435 .
## targetref:qd 4.413e-01 1.254e+01 1.238e+05 0.035 0.971935
## targetref:qe -2.321e+01 9.511e+00 1.239e+05 -2.441 0.014662 *
## targetref:qf 1.801e+01 1.210e+01 1.238e+05 1.488 0.136766
## targetref:det -1.478e+01 1.084e+01 1.239e+05 -1.363 0.172784
## qb:det 5.915e+01 1.678e+01 1.219e+05 3.526 0.000423 ***
## qd:det 3.648e+01 2.225e+02 1.184e+05 0.164 0.869736
## targetref:qb:det -5.198e+01 2.448e+01 1.233e+05 -2.123 0.033761 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 7 columns / coefficients
## Some predictor variables are on very different scales: consider rescaling3.2.4 plots
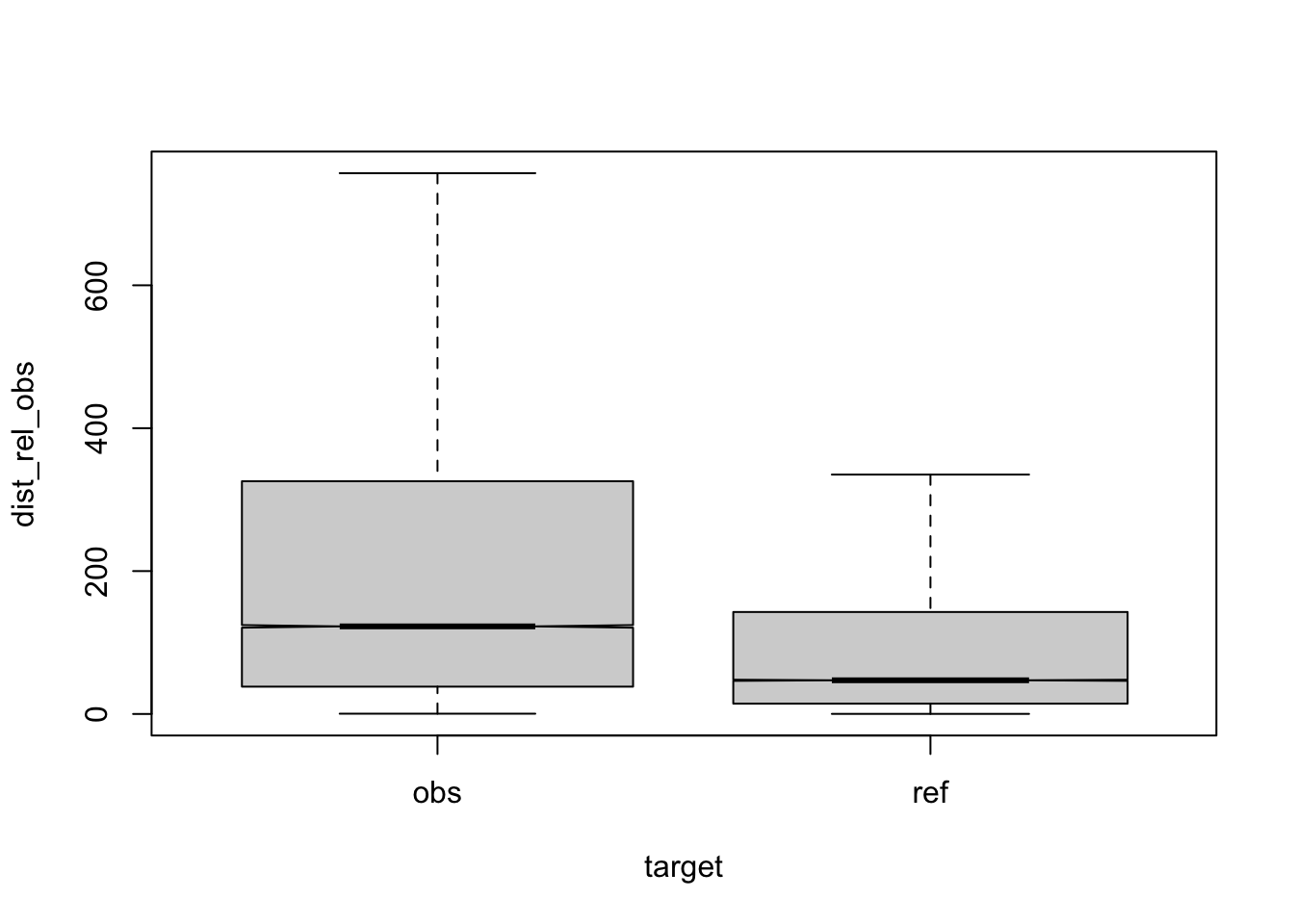
Figure 3.1: compare distances by corpus, normalised to obs, distance ceiling = outliers removed
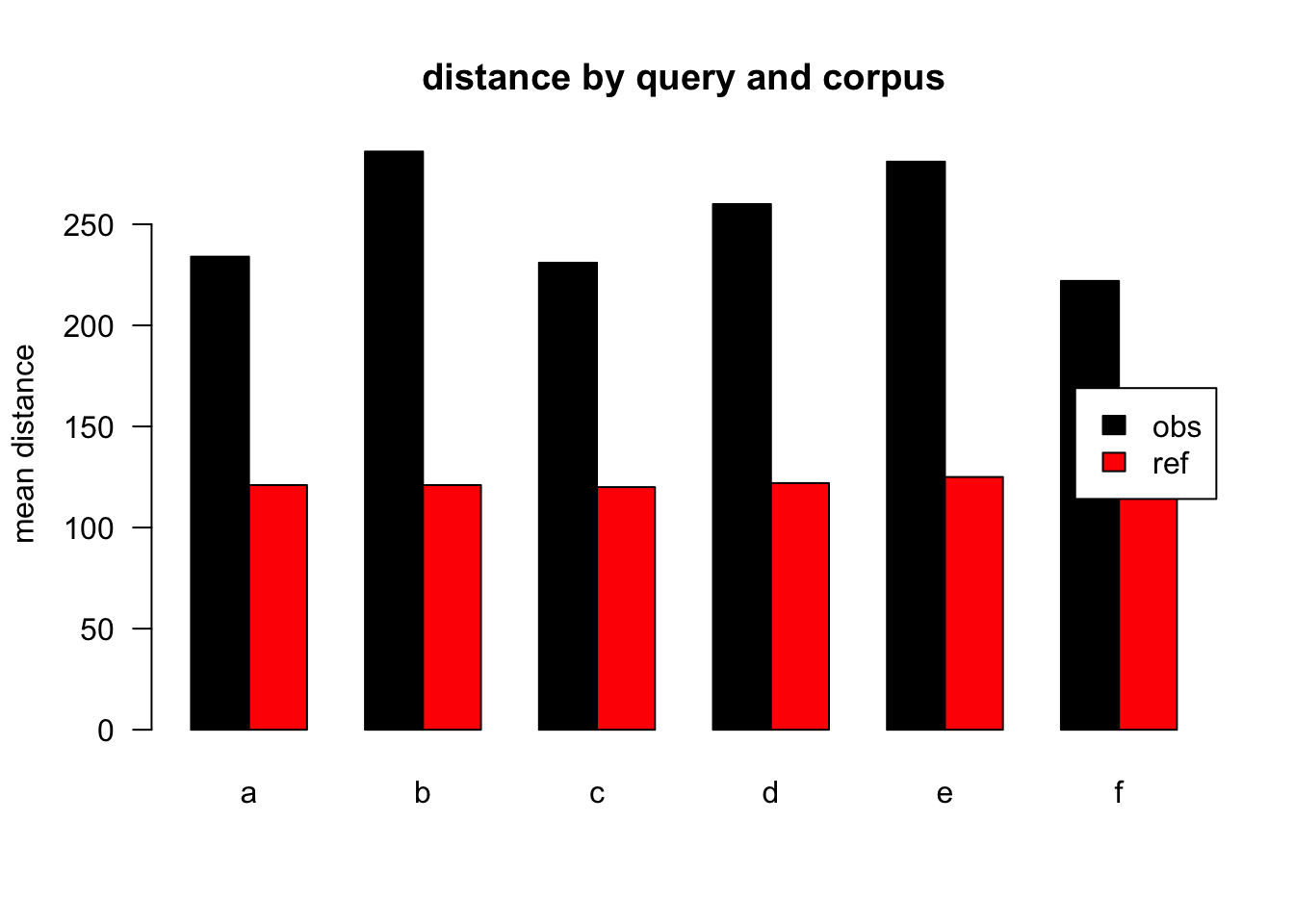
Figure 3.2: mean distances over query/corpus, normalised to obs, distance ceiling = outliers removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 42836 | 234 | 117 |
| ref | a | 58615 | 121 | 47 |
| obs | b | 2116 | 286 | 165 |
| ref | b | 1130 | 121 | 44 |
| obs | c | 5770 | 231 | 114 |
| ref | c | 1274 | 120 | 48 |
| obs | d | 5654 | 260 | 144 |
| ref | d | 1525 | 122 | 49 |
| obs | e | 3911 | 281 | 147 |
| ref | e | 671 | 125 | 45 |
| obs | f | 2311 | 222 | 133 |
| ref | f | 413 | 116 | 47 |
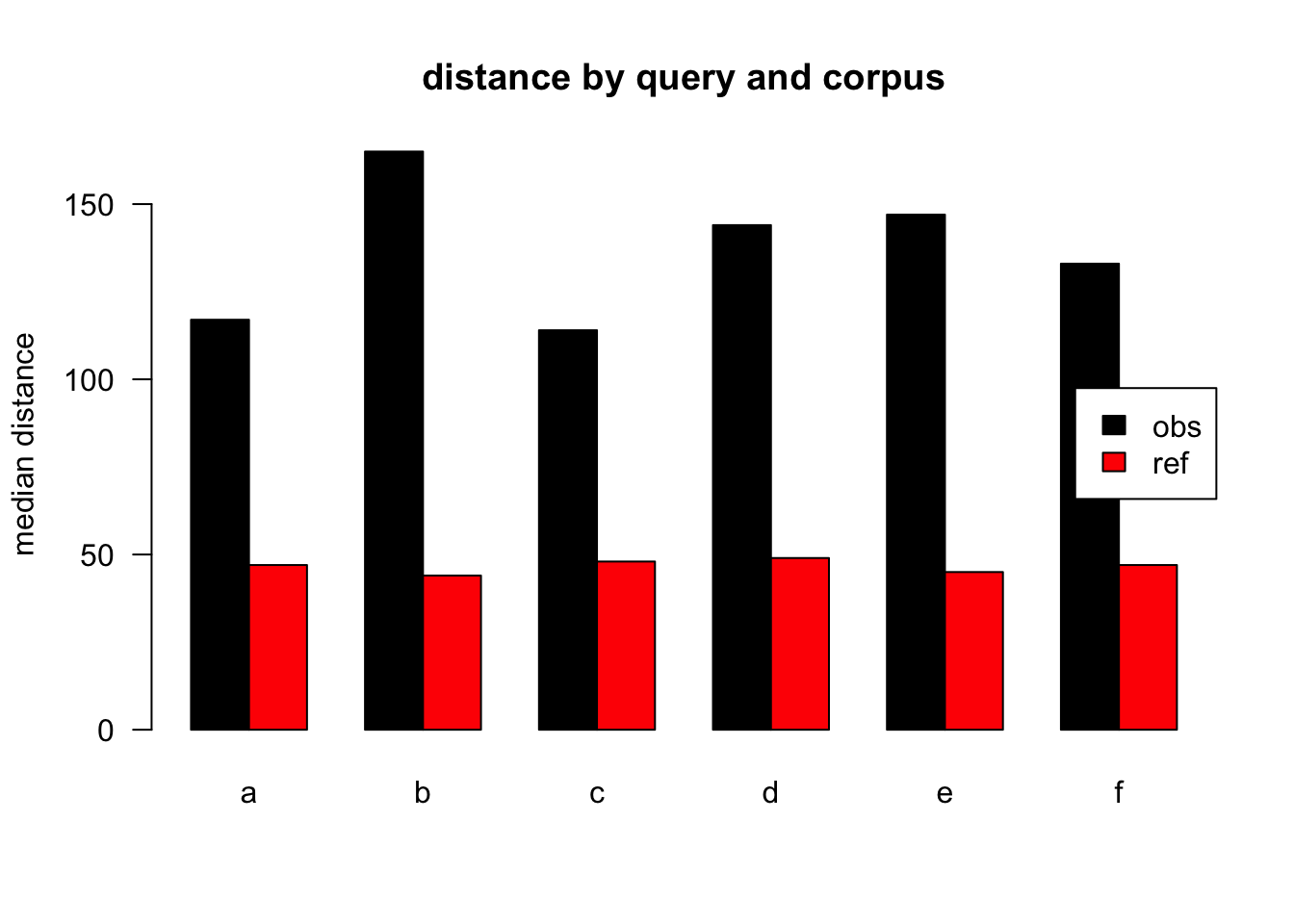
Figure 3.3: median distances over query/corpus, normalised to obs, distance ceiling = outliers removed
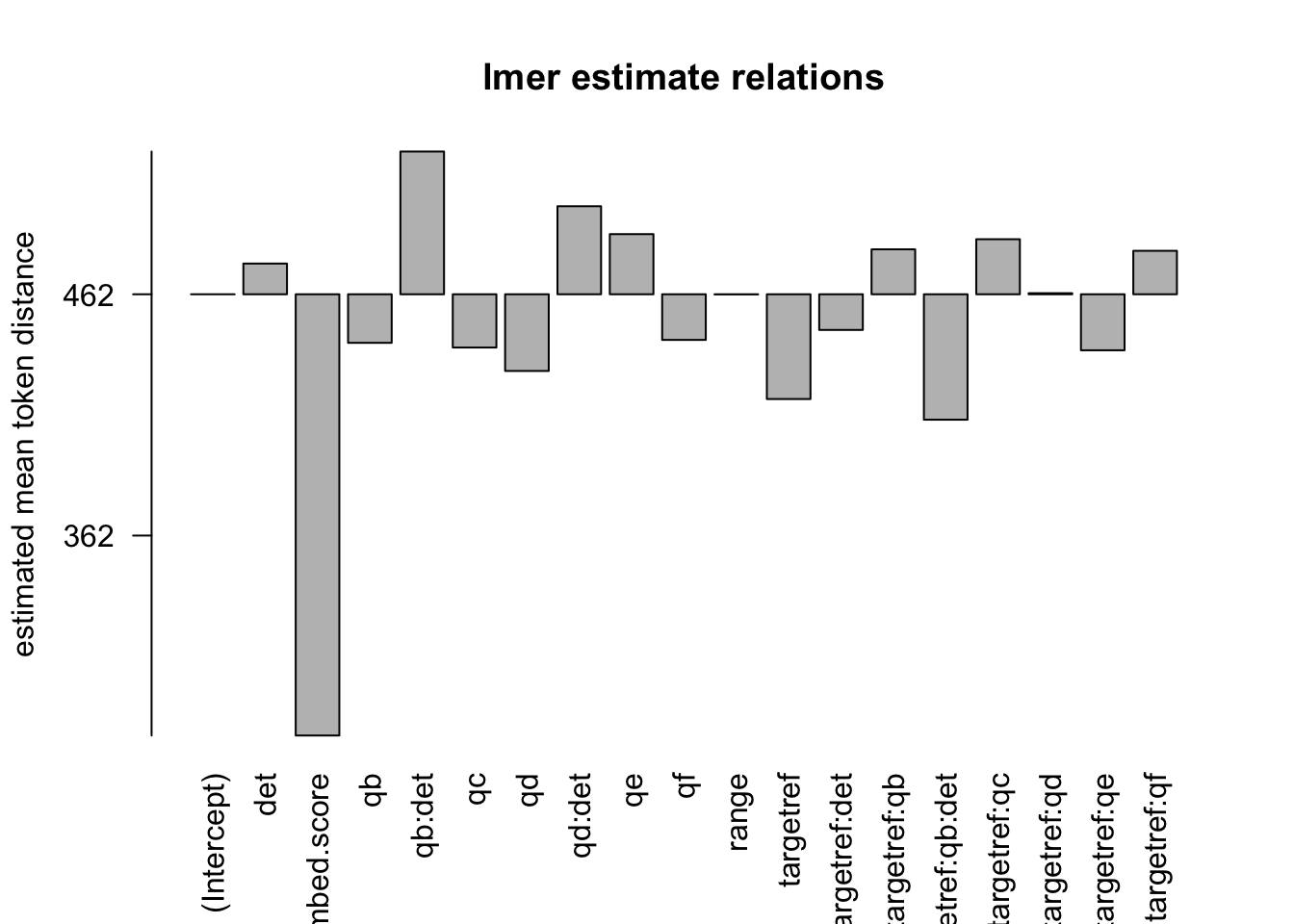
Figure 3.4: distances relation, normalised to obs, distance ceiling = outliers removed
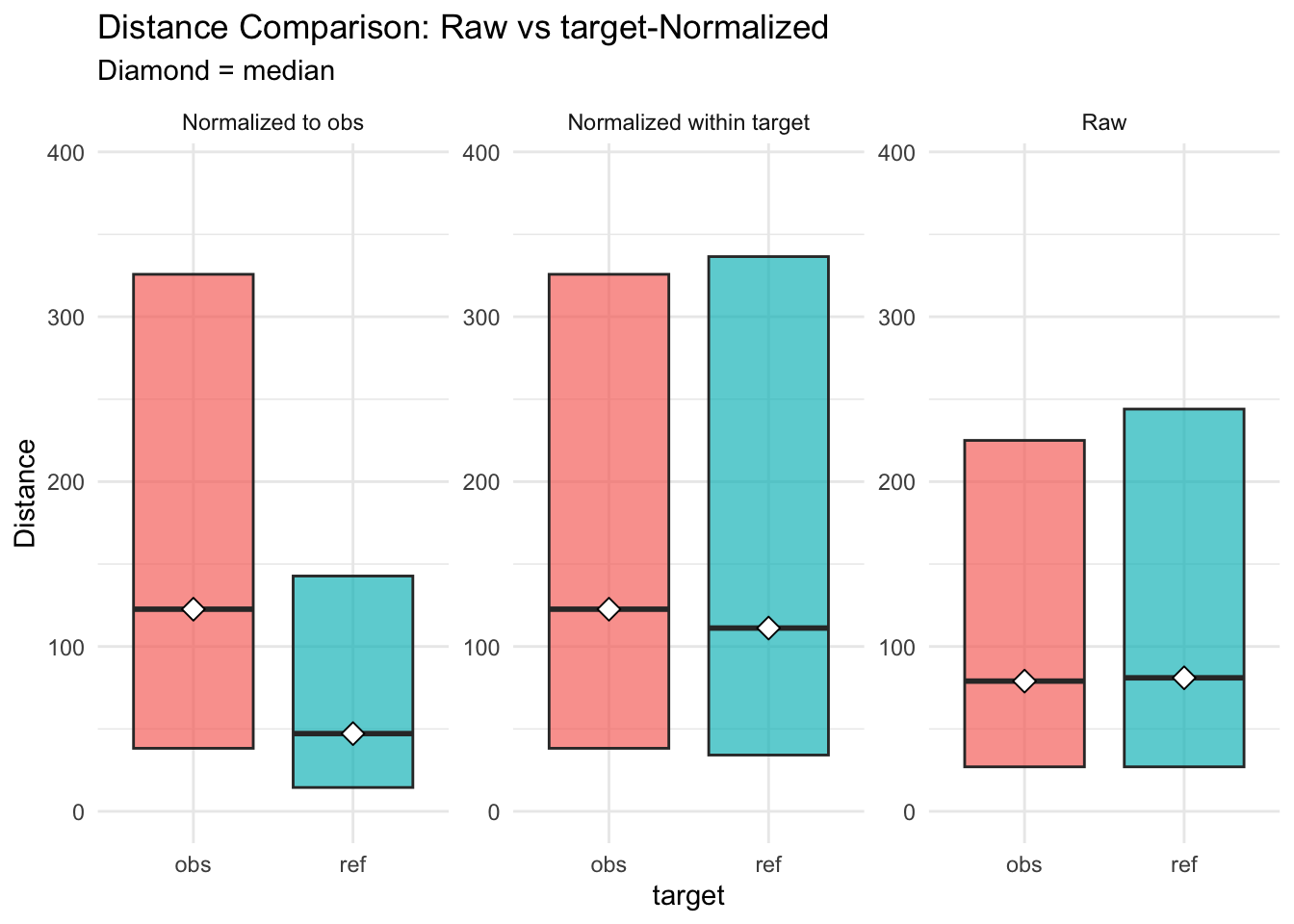
Figure 3.5: distances normalised vs. raw
3.3 evaluation model: 2
3.3.2 parameter setting
## value
## norm_target
## det.t TRUE
## limit FALSE
## author TRUE
## url TRUE
## embed1 TRUE
## embed2 f
## range1 TRUE
## range2 f
## rel FALSE
## lme FALSE
## lemma FALSE3.3.3 anova analysis
3.3.3.1 anova plain
formula: [dist ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 1.1152e+11 1.1152e+11 268.8154 < 2e-16 ***
## q 5 9.8792e+08 1.9758e+08 0.4763 0.79425
## det 1 4.1537e+08 4.1537e+08 1.0012 0.31702
## target:q 5 2.3050e+09 4.6101e+08 1.1112 0.35184
## target:det 1 2.7199e+09 2.7199e+09 6.5561 0.01045 *
## q:det 2 2.4028e+08 1.2014e+08 0.2896 0.74857
## target:q:det 1 7.0024e+06 7.0024e+06 0.0169 0.89663
## Residuals 142304 5.9037e+13 4.1487e+08
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.3.3.2 anova of linear regression model
[anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 1.2717e+09 1.2717e+09 1 3751 5.5781 0.01824 *
## q 6.3534e+08 1.2707e+08 5 137654 0.5574 0.73281
## det 7.3359e+05 7.3359e+05 1 133172 0.0032 0.95476
## range 2.8637e+07 2.8637e+07 1 2113 0.1256 0.72306
## embed.score 2.7199e+10 2.7199e+10 1 141732 119.3005 < 2e-16 ***
## target:q 3.0753e+09 6.1507e+08 5 138840 2.6979 0.01920 *
## target:det 8.1028e+08 8.1028e+08 1 138434 3.5541 0.05940 .
## q:det 4.8717e+08 2.4358e+08 2 135770 1.0684 0.34355
## target:q:det 2.4585e+06 2.4585e+06 1 138496 0.0108 0.91729
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.3.3.3 linear regression coefficients
formula: [dist ~ target*q*det+(1|aut_id)+range+(embed.score)+(1|url_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 3153644
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -23.760 -0.034 -0.006 0.025 55.672
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 28985985 5384
## url_id (Intercept) 98381104 9919
## Residual 227983636 15099
## Number of obs: 142321, groups: aut_id, 8395; url_id, 2145
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.873e+03 4.211e+02 8.594e+03 6.823 9.53e-12 ***
## targetref 1.341e+03 6.536e+02 2.412e+03 2.051 0.0404 *
## qb 6.895e+01 1.008e+03 1.363e+05 0.068 0.9454
## qc -6.307e+02 3.622e+02 1.372e+05 -1.741 0.0816 .
## qd -1.993e+03 1.522e+04 1.332e+05 -0.131 0.8958
## qe -1.006e+02 2.520e+02 1.385e+05 -0.399 0.6899
## qf -1.355e+02 3.218e+02 1.384e+05 -0.421 0.6737
## det 7.031e+02 3.145e+02 1.375e+05 2.236 0.0254 *
## range 6.798e-02 1.918e-01 2.113e+03 0.354 0.7231
## embed.score -5.793e+03 5.304e+02 1.417e+05 -10.922 < 2e-16 ***
## targetref:qb 6.675e+02 1.124e+03 1.371e+05 0.594 0.5527
## targetref:qc 3.752e+01 8.128e+02 1.395e+05 0.046 0.9632
## targetref:qd 2.022e+03 7.989e+02 1.395e+05 2.531 0.0114 *
## targetref:qe 2.269e+02 6.042e+02 1.395e+05 0.376 0.7073
## targetref:qf 3.210e+02 7.643e+02 1.393e+05 0.420 0.6745
## targetref:det -1.416e+03 6.890e+02 1.397e+05 -2.055 0.0398 *
## qb:det -1.077e+03 1.107e+03 1.364e+05 -0.973 0.3304
## qd:det 1.039e+03 1.521e+04 1.332e+05 0.068 0.9456
## targetref:qb:det -1.651e+02 1.590e+03 1.385e+05 -0.104 0.9173
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 7 columns / coefficients
## Some predictor variables are on very different scales: consider rescaling3.3.4 plots
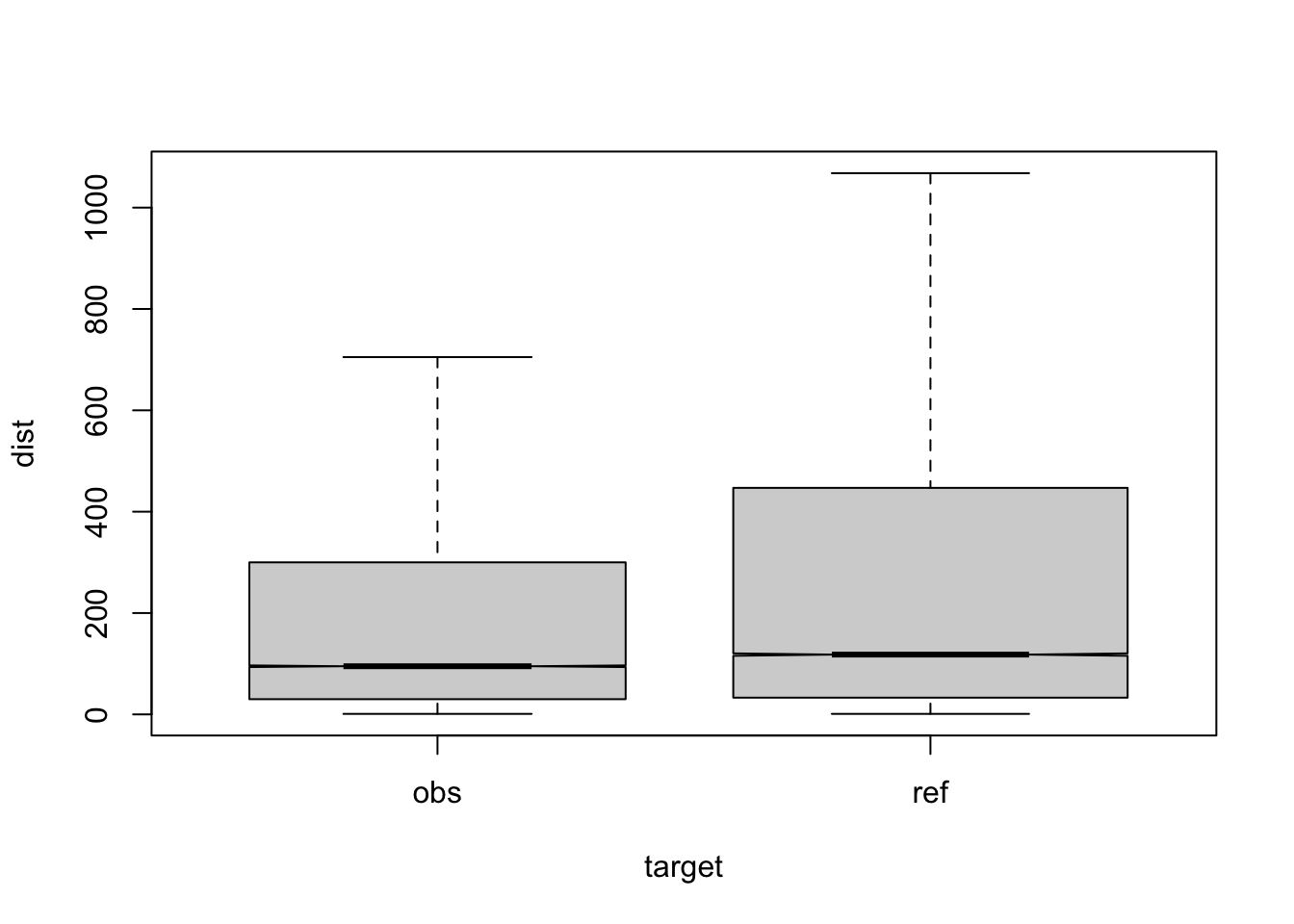
Figure 3.6: compare distances by corpus, not normalised, distance ceiling =outliers not removed
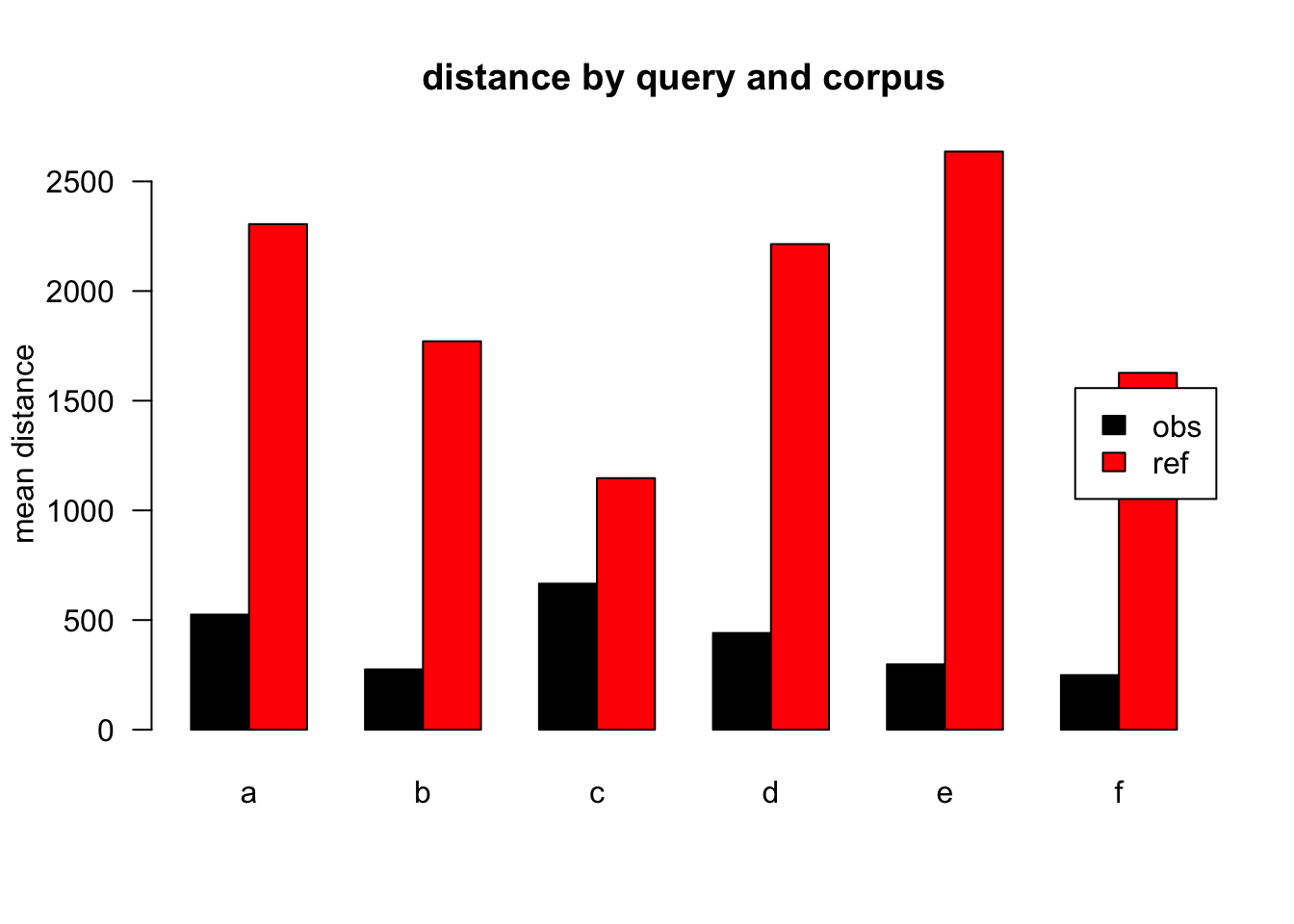
Figure 3.7: mean distances over query/corpus, not normalised, distance ceiling =outliers not removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 46318 | 525 | 92 |
| ref | a | 68618 | 2305 | 118 |
| obs | b | 2287 | 275 | 109 |
| ref | b | 1315 | 1771 | 111 |
| obs | c | 6253 | 666 | 89 |
| ref | c | 1504 | 1147 | 119 |
| obs | d | 6171 | 441 | 105 |
| ref | d | 1765 | 2214 | 124 |
| obs | e | 4278 | 298 | 109 |
| ref | e | 795 | 2636 | 116 |
| obs | f | 2520 | 249 | 77 |
| ref | f | 497 | 1627 | 124 |
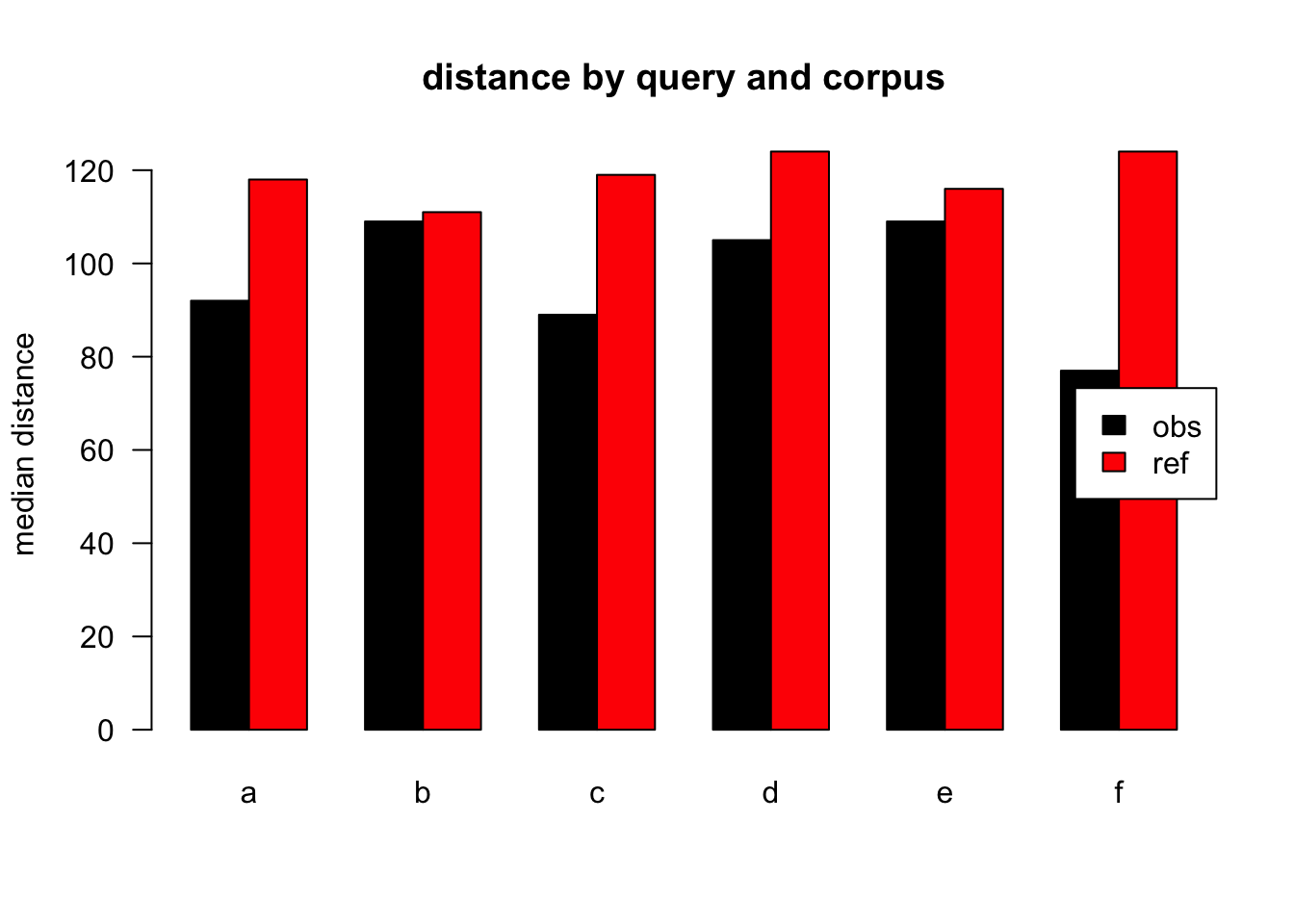
Figure 3.8: median distances over query/corpus, not normalised, distance ceiling =outliers not removed
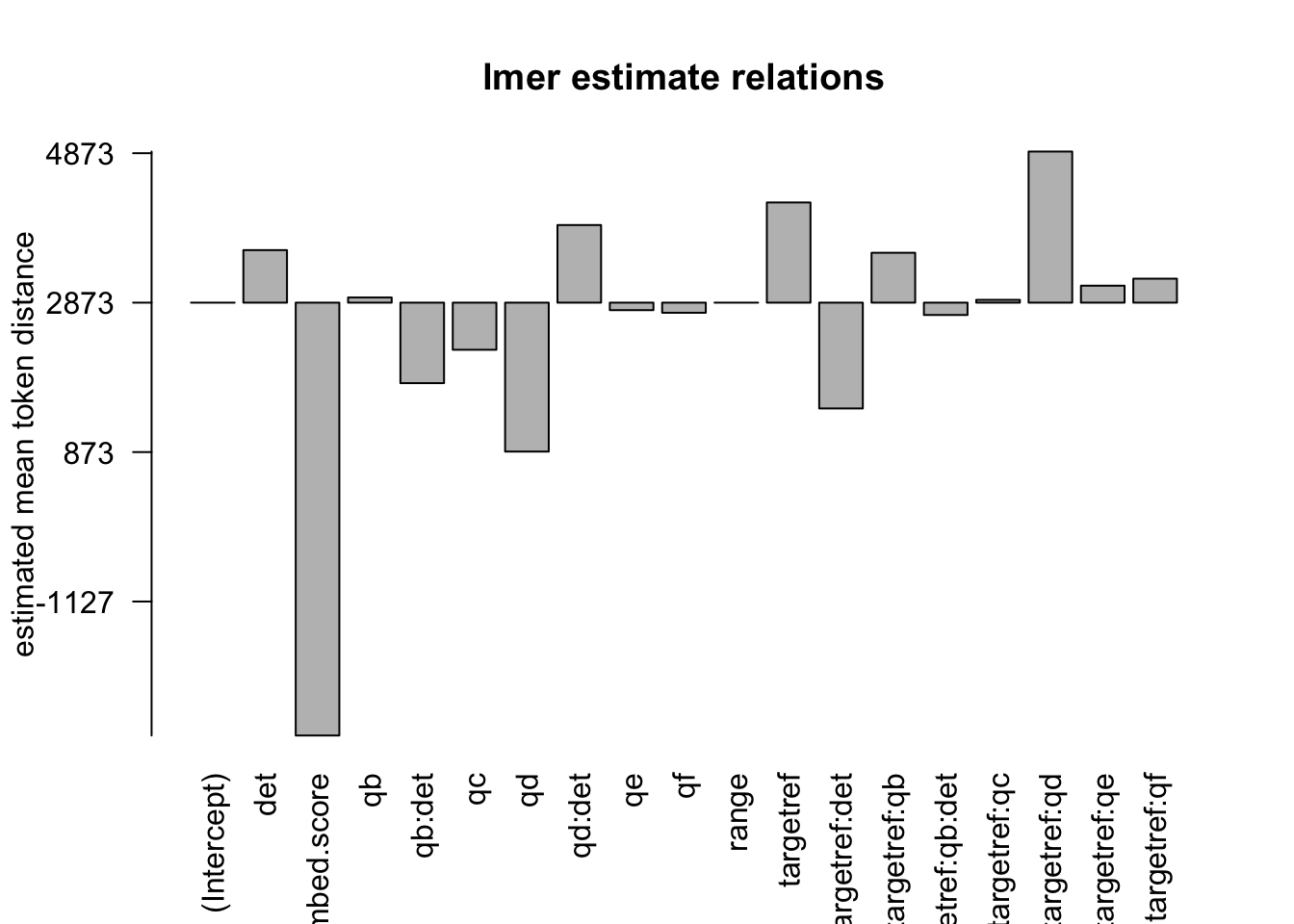
Figure 3.9: distances relation, not normalised, distance ceiling =outliers not removed
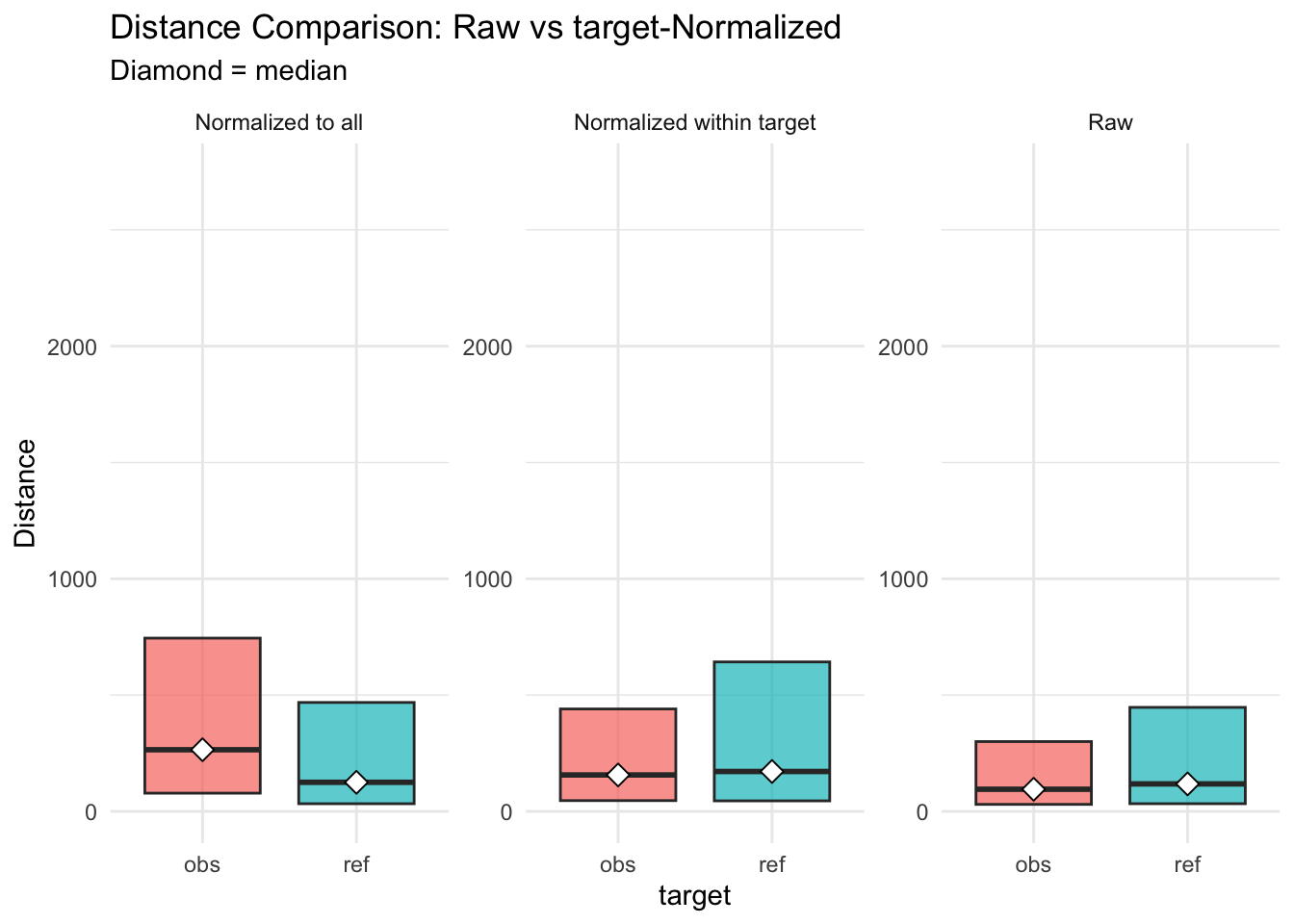
Figure 3.10: distances normalised vs. raw
3.4 evaluation model: 3
3.4.2 parameter setting
## value
## norm_target _rel_all
## det.t TRUE
## limit TRUE
## author TRUE
## url TRUE
## embed1 TRUE
## embed2 f
## range1 TRUE
## range2 f
## rel TRUE
## lme FALSE
## lemma FALSE3.4.3 anova analysis
3.4.3.1 anova plain
formula: [dist_rel_all ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 1.2830e+09 1283010757 7336.4625 < 2.2e-16 ***
## q 5 3.4949e+07 6989793 39.9688 < 2.2e-16 ***
## det 1 4.6410e+06 4641007 26.5380 2.588e-07 ***
## target:q 5 7.7932e+06 1558646 8.9126 1.786e-08 ***
## target:det 1 7.1283e+05 712833 4.0761 0.043496 *
## q:det 2 2.5680e+06 1283981 7.3420 0.000648 ***
## target:q:det 1 2.0345e+06 2034482 11.6335 0.000648 ***
## Residuals 126209 2.2072e+10 174881
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.4.3.2 anova of linear regression model
[anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 3245706 3245706 1 3519 23.4567 1.333e-06 ***
## q 2091953 418391 5 122421 3.0237 0.0098706 **
## det 34508 34508 1 118425 0.2494 0.6175055
## range 142964301 142964301 1 1025 1033.2042 < 2.2e-16 ***
## embed.score 71204325 71204325 1 122690 514.5942 < 2.2e-16 ***
## target:q 2202162 440432 5 123486 3.1830 0.0070933 **
## target:det 1534830 1534830 1 123325 11.0922 0.0008672 ***
## q:det 1019818 509909 2 120804 3.6851 0.0250971 *
## target:q:det 623611 623611 1 123315 4.5068 0.0337615 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.4.3.3 linear regression coefficients
formula: [dist_rel_all ~ target*q*det+(1|aut_id)+range+(embed.score)+(1|url_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 1859224
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.8643 -0.5282 -0.1721 0.2469 6.9244
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 8101 90.01
## url_id (Intercept) 23223 152.39
## Residual 138370 371.98
## Number of obs: 126226, groups: aut_id, 8238; url_id, 2145
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 7.789e+02 8.688e+00 8.969e+03 89.651 < 2e-16 ***
## targetref -7.312e+01 1.061e+01 1.300e+03 -6.893 8.50e-12 ***
## qb -3.390e+01 2.572e+01 1.218e+05 -1.318 0.187483
## qc -3.717e+01 9.261e+00 1.226e+05 -4.014 5.98e-05 ***
## qd -5.353e+01 3.748e+02 1.184e+05 -0.143 0.886426
## qe 4.198e+01 6.460e+00 1.247e+05 6.498 8.14e-11 ***
## qf -3.185e+01 8.240e+00 1.244e+05 -3.866 0.000111 ***
## det 2.144e+01 8.041e+00 1.229e+05 2.667 0.007662 **
## range -9.786e-02 3.044e-03 1.025e+03 -32.143 < 2e-16 ***
## embed.score -3.080e+02 1.358e+01 1.227e+05 -22.685 < 2e-16 ***
## targetref:qb 3.136e+01 2.894e+01 1.225e+05 1.083 0.278599
## targetref:qc 3.842e+01 2.154e+01 1.237e+05 1.784 0.074435 .
## targetref:qd 7.432e-01 2.113e+01 1.238e+05 0.035 0.971935
## targetref:qe -3.910e+01 1.602e+01 1.239e+05 -2.441 0.014662 *
## targetref:qf 3.033e+01 2.039e+01 1.238e+05 1.488 0.136766
## targetref:det -2.490e+01 1.826e+01 1.239e+05 -1.363 0.172784
## qb:det 9.962e+01 2.826e+01 1.219e+05 3.526 0.000423 ***
## qd:det 6.144e+01 3.747e+02 1.184e+05 0.164 0.869736
## targetref:qb:det -8.754e+01 4.124e+01 1.233e+05 -2.123 0.033761 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 7 columns / coefficients
## Some predictor variables are on very different scales: consider rescaling3.4.4 plots
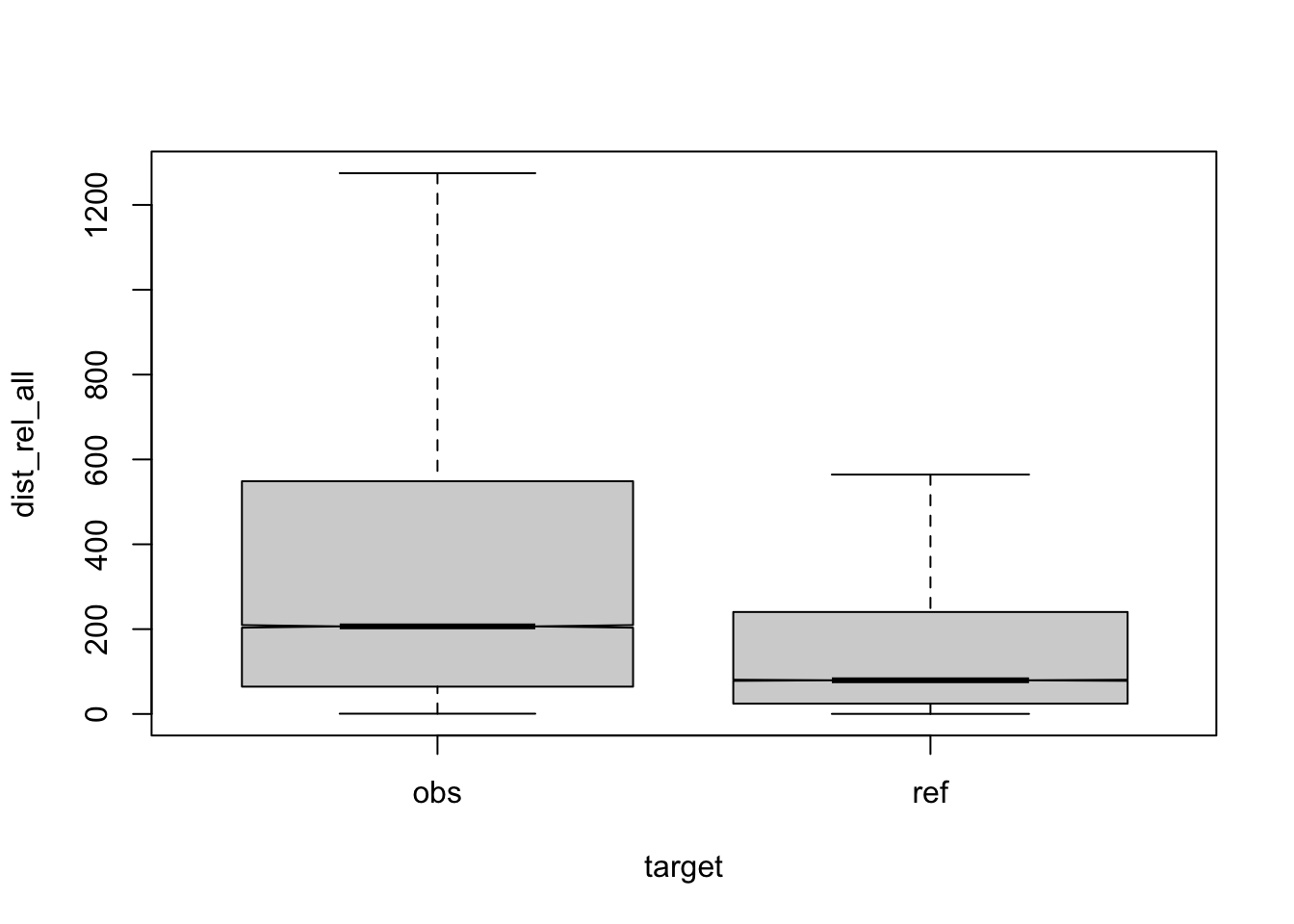
Figure 3.11: compare distances by corpus, normalised to all, distance ceiling = outliers removed
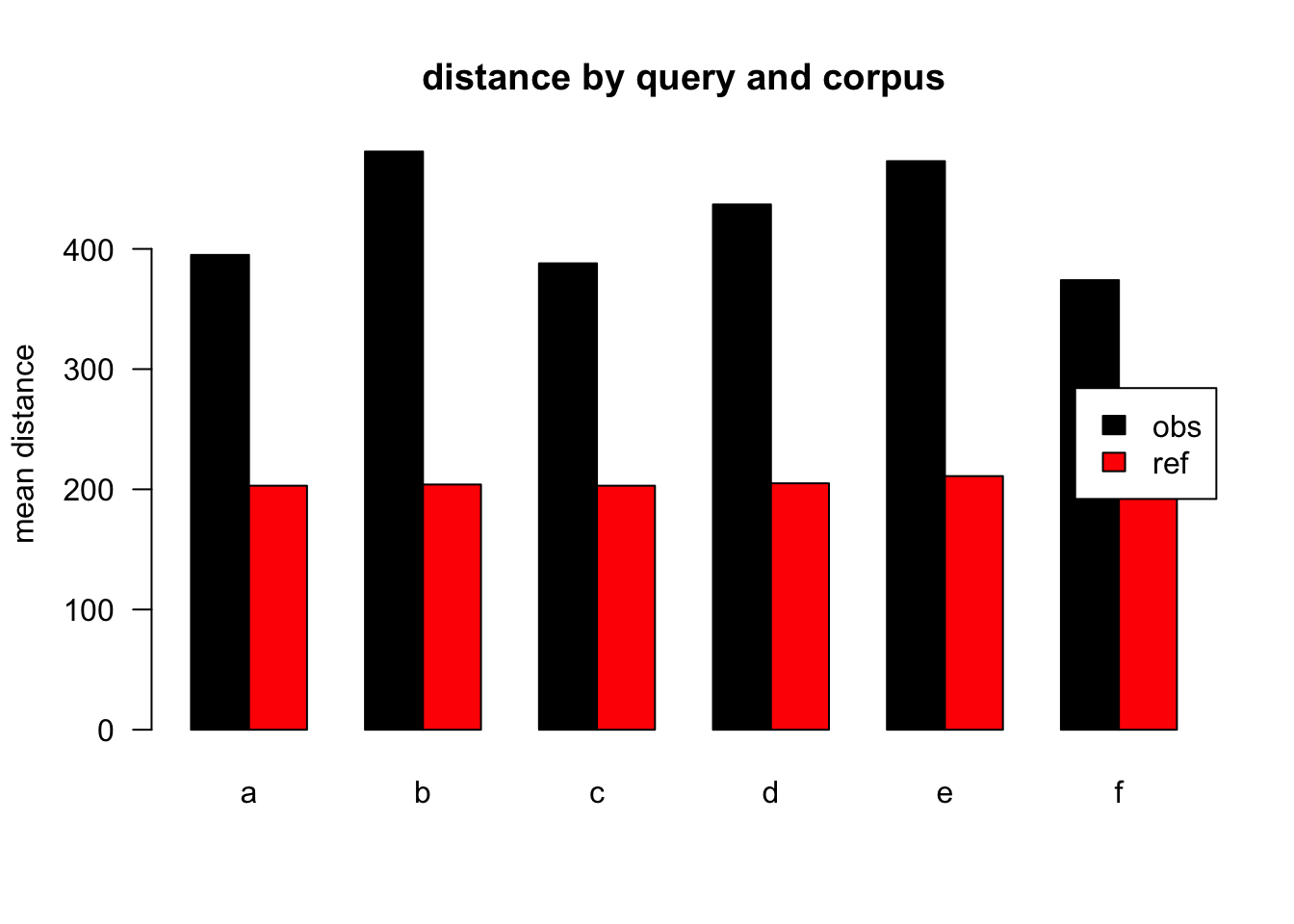
Figure 3.12: mean distances over query/corpus, normalised to all, distance ceiling = outliers removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 42836 | 395 | 196 |
| ref | a | 58615 | 203 | 79 |
| obs | b | 2116 | 481 | 279 |
| ref | b | 1130 | 204 | 75 |
| obs | c | 5770 | 388 | 191 |
| ref | c | 1274 | 203 | 80 |
| obs | d | 5654 | 437 | 243 |
| ref | d | 1525 | 205 | 83 |
| obs | e | 3911 | 473 | 248 |
| ref | e | 671 | 211 | 75 |
| obs | f | 2311 | 374 | 224 |
| ref | f | 413 | 195 | 79 |
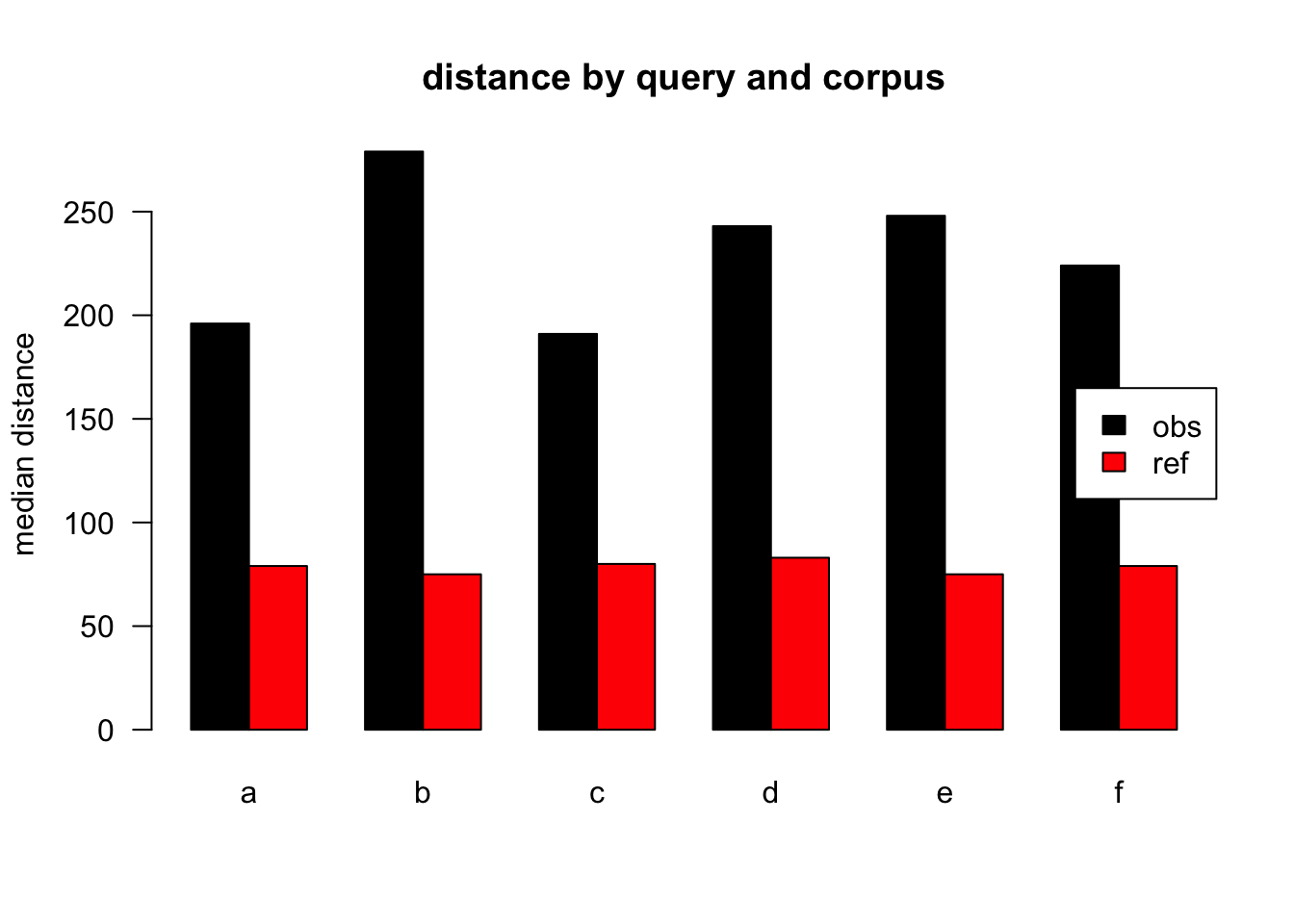
Figure 3.13: median distances over query/corpus, normalised to all, distance ceiling = outliers removed
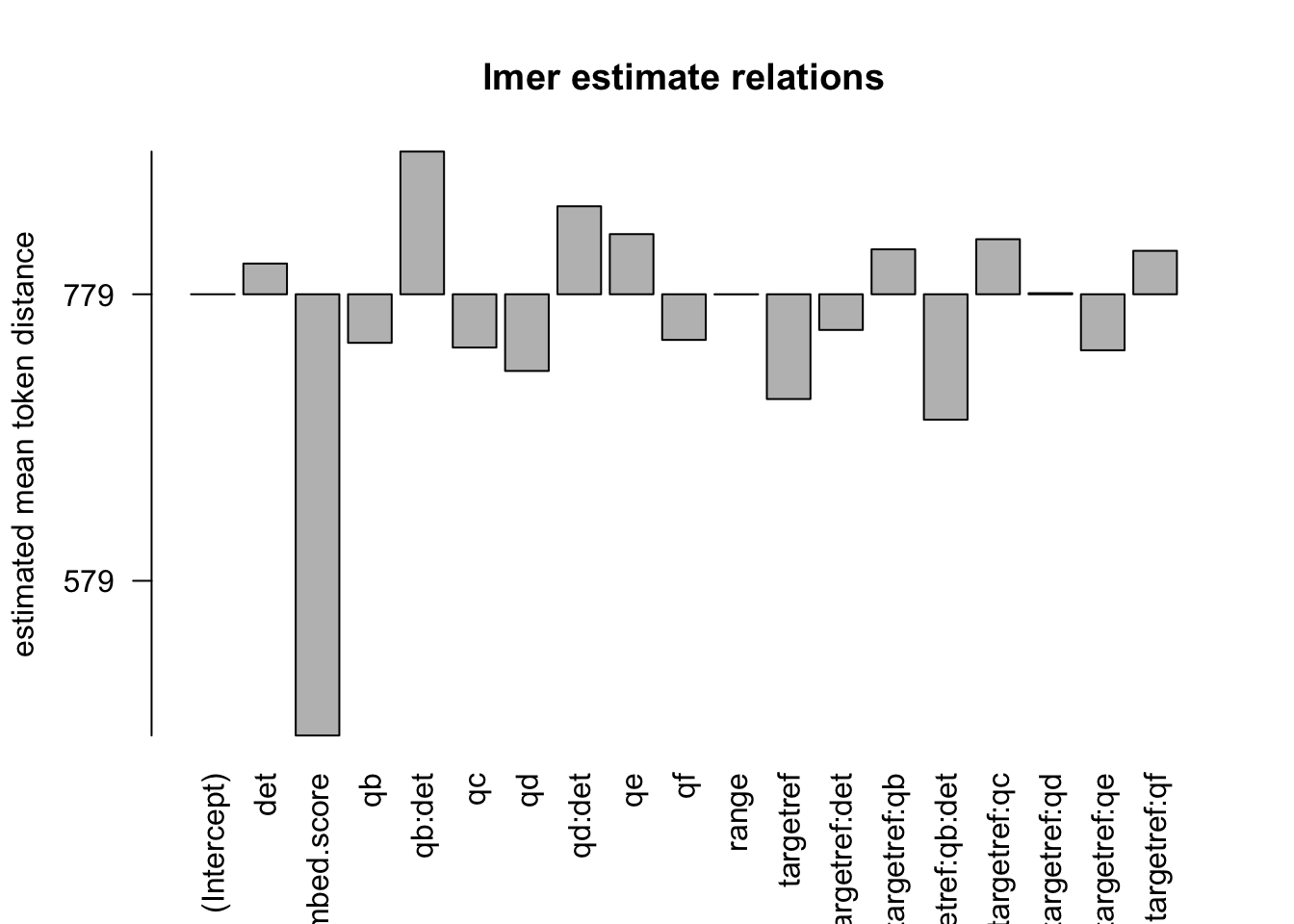
Figure 3.14: distances relation, normalised to all, distance ceiling = outliers removed
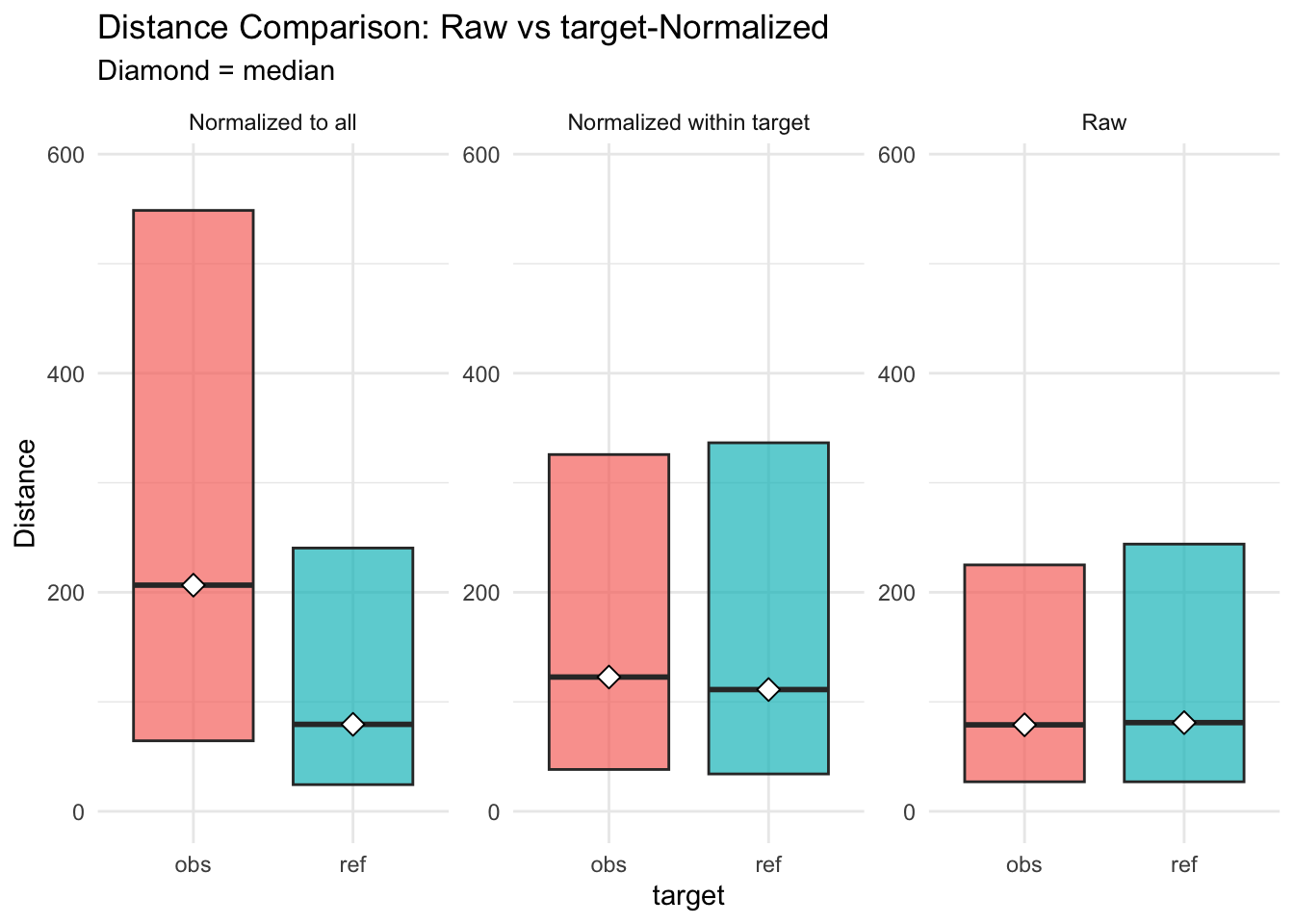
Figure 3.15: distances normalised vs. raw
3.5 evaluation model: 4
3.5.2 parameter setting
## value
## norm_target _rel_ref
## det.t TRUE
## limit TRUE
## author TRUE
## url TRUE
## embed1 TRUE
## embed2 f
## range1 TRUE
## range2 f
## rel TRUE
## lme FALSE
## lemma FALSE3.5.3 anova analysis
3.5.3.1 anova plain
formula: [dist_rel_ref ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 2.5135e+09 2513546743 7336.4625 < 2.2e-16 ***
## q 5 6.8469e+07 13693706 39.9688 < 2.2e-16 ***
## det 1 9.0922e+06 9092198 26.5380 2.588e-07 ***
## target:q 5 1.5268e+07 3053543 8.9126 1.786e-08 ***
## target:det 1 1.3965e+06 1396511 4.0761 0.043496 *
## q:det 2 5.0309e+06 2515448 7.3420 0.000648 ***
## target:q:det 1 3.9858e+06 3985754 11.6335 0.000648 ***
## Residuals 126209 4.3240e+10 342610
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.5.3.2 anova of linear regression model
[anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 6358663 6358663 1 3519 23.4567 1.333e-06 ***
## q 4098347 819669 5 122421 3.0237 0.0098706 **
## det 67605 67605 1 118425 0.2494 0.6175055
## range 280081403 280081403 1 1025 1033.2042 < 2.2e-16 ***
## embed.score 139496414 139496414 1 122690 514.5942 < 2.2e-16 ***
## target:q 4314256 862851 5 123486 3.1830 0.0070933 **
## target:det 3006886 3006886 1 123325 11.0922 0.0008672 ***
## q:det 1997926 998963 2 120804 3.6851 0.0250971 *
## target:q:det 1221717 1221717 1 123315 4.5068 0.0337615 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.5.3.3 linear regression coefficients
formula: [dist_rel_ref ~ target*q*det+(1|aut_id)+range+(embed.score)+(1|url_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 1944096
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.8643 -0.5282 -0.1721 0.2469 6.9244
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 15871 126.0
## url_id (Intercept) 45496 213.3
## Residual 271080 520.7
## Number of obs: 126226, groups: aut_id, 8238; url_id, 2145
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 1.090e+03 1.216e+01 8.969e+03 89.651 < 2e-16 ***
## targetref -1.024e+02 1.485e+01 1.300e+03 -6.893 8.50e-12 ***
## qb -4.744e+01 3.600e+01 1.218e+05 -1.318 0.187483
## qc -5.203e+01 1.296e+01 1.226e+05 -4.014 5.98e-05 ***
## qd -7.492e+01 5.246e+02 1.184e+05 -0.143 0.886426
## qe 5.876e+01 9.042e+00 1.247e+05 6.498 8.14e-11 ***
## qf -4.458e+01 1.153e+01 1.244e+05 -3.866 0.000111 ***
## det 3.001e+01 1.125e+01 1.229e+05 2.667 0.007662 **
## range -1.370e-01 4.261e-03 1.025e+03 -32.143 < 2e-16 ***
## embed.score -4.311e+02 1.900e+01 1.227e+05 -22.685 < 2e-16 ***
## targetref:qb 4.389e+01 4.051e+01 1.225e+05 1.083 0.278599
## targetref:qc 5.378e+01 3.015e+01 1.237e+05 1.784 0.074435 .
## targetref:qd 1.040e+00 2.957e+01 1.238e+05 0.035 0.971935
## targetref:qe -5.472e+01 2.242e+01 1.239e+05 -2.441 0.014662 *
## targetref:qf 4.246e+01 2.853e+01 1.238e+05 1.488 0.136766
## targetref:det -3.485e+01 2.556e+01 1.239e+05 -1.363 0.172784
## qb:det 1.394e+02 3.955e+01 1.219e+05 3.526 0.000423 ***
## qd:det 8.600e+01 5.244e+02 1.184e+05 0.164 0.869736
## targetref:qb:det -1.225e+02 5.772e+01 1.233e+05 -2.123 0.033761 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 7 columns / coefficients
## Some predictor variables are on very different scales: consider rescaling3.5.4 plots
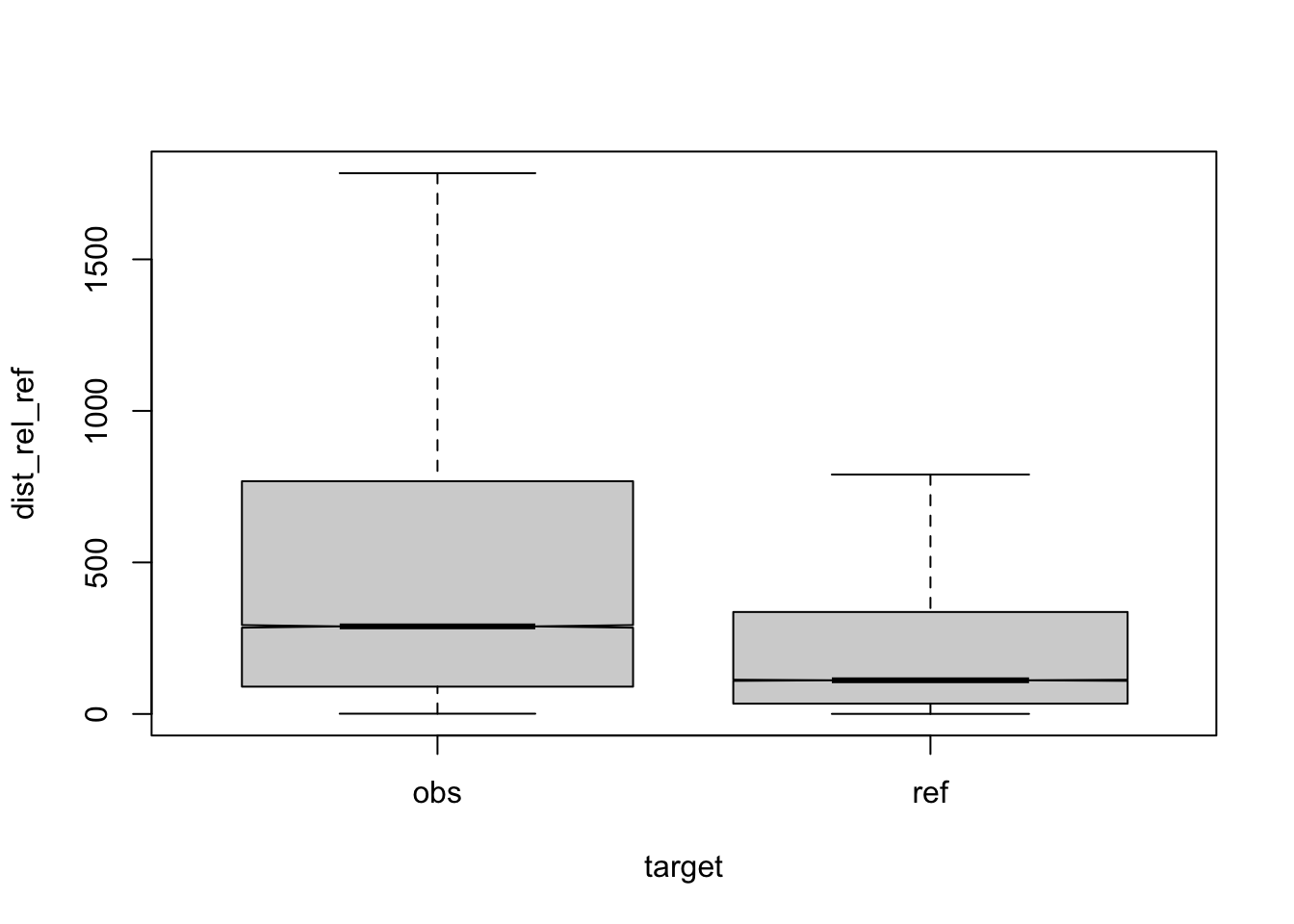
Figure 3.16: compare distances by corpus, normalised to ref, distance ceiling = outliers removed
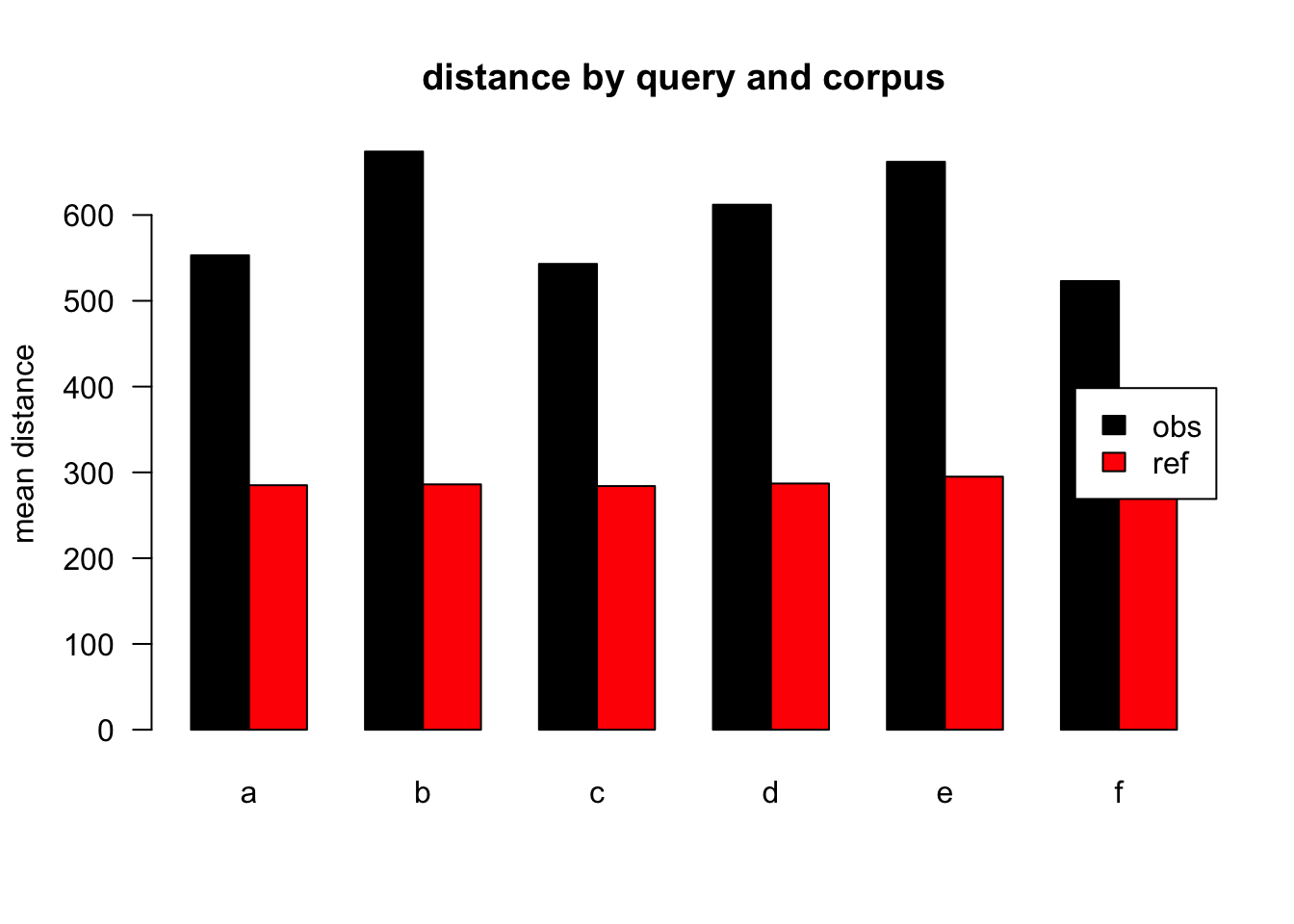
Figure 3.17: mean distances over query/corpus, normalised to ref, distance ceiling = outliers removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 42836 | 553 | 275 |
| ref | a | 58615 | 285 | 111 |
| obs | b | 2116 | 674 | 390 |
| ref | b | 1130 | 286 | 104 |
| obs | c | 5770 | 543 | 268 |
| ref | c | 1274 | 284 | 112 |
| obs | d | 5654 | 612 | 340 |
| ref | d | 1525 | 287 | 116 |
| obs | e | 3911 | 662 | 347 |
| ref | e | 671 | 295 | 105 |
| obs | f | 2311 | 523 | 313 |
| ref | f | 413 | 273 | 111 |
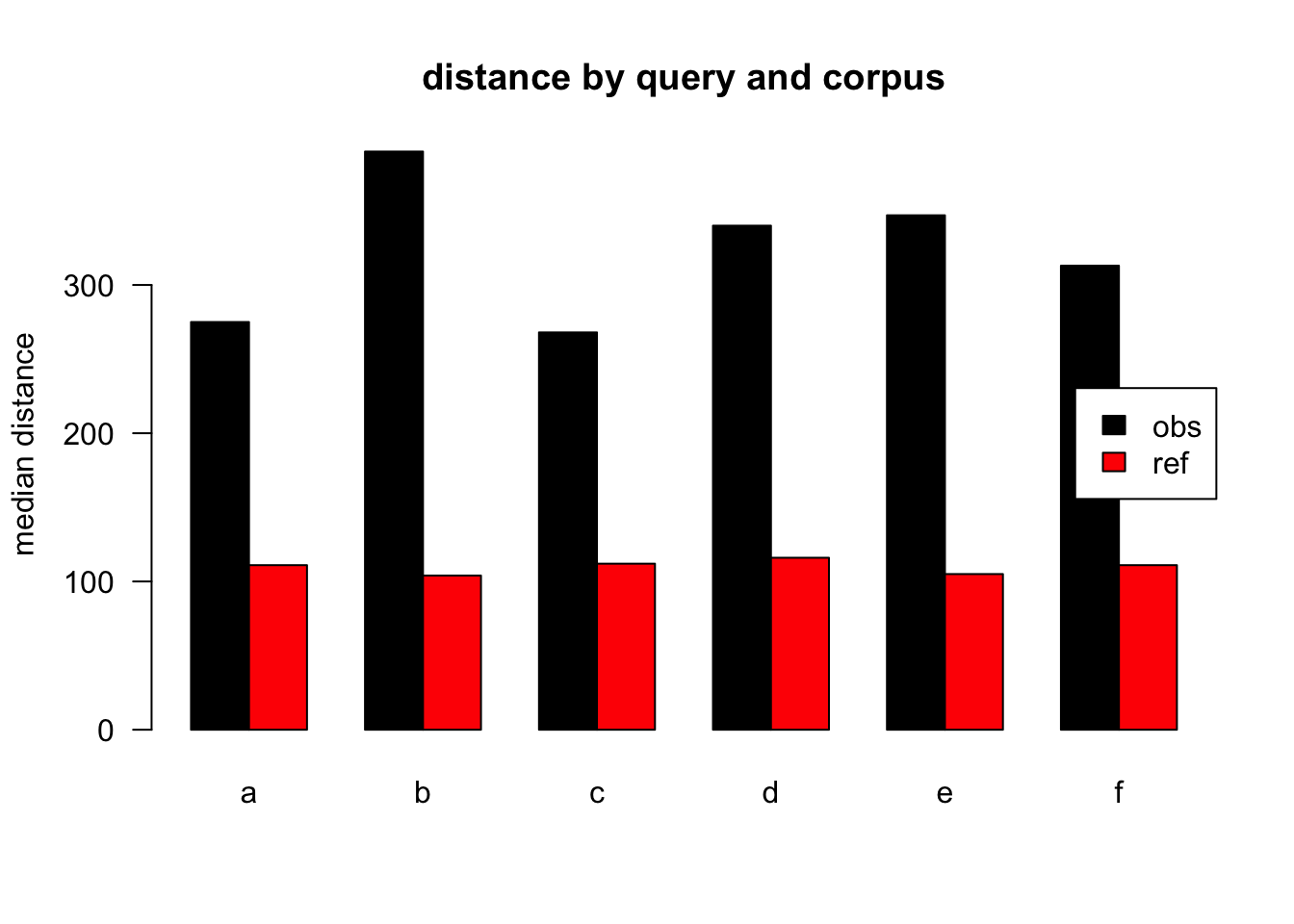
Figure 3.18: median distances over query/corpus, normalised to ref, distance ceiling = outliers removed
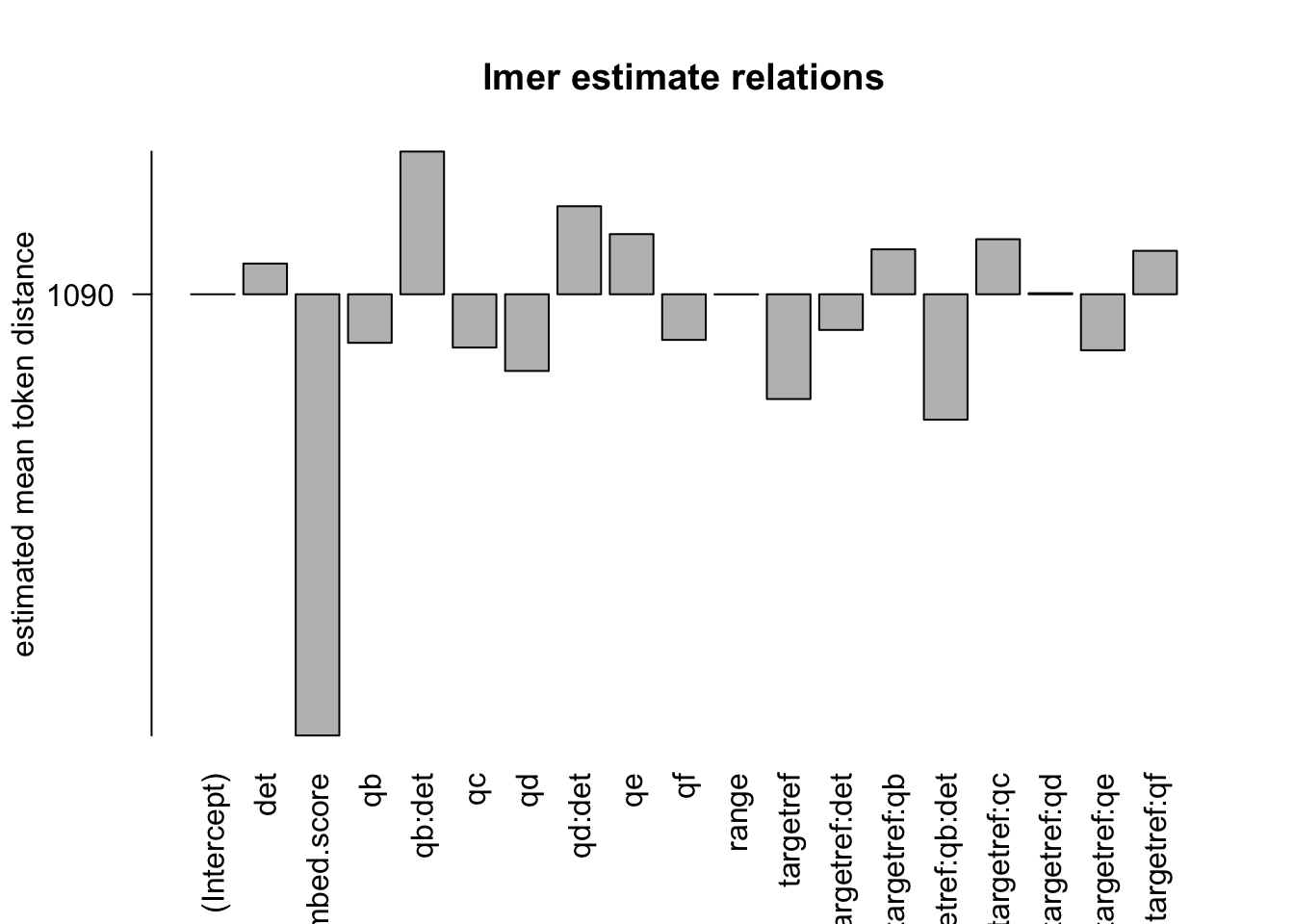
Figure 3.19: distances relation, normalised to ref, distance ceiling = outliers removed
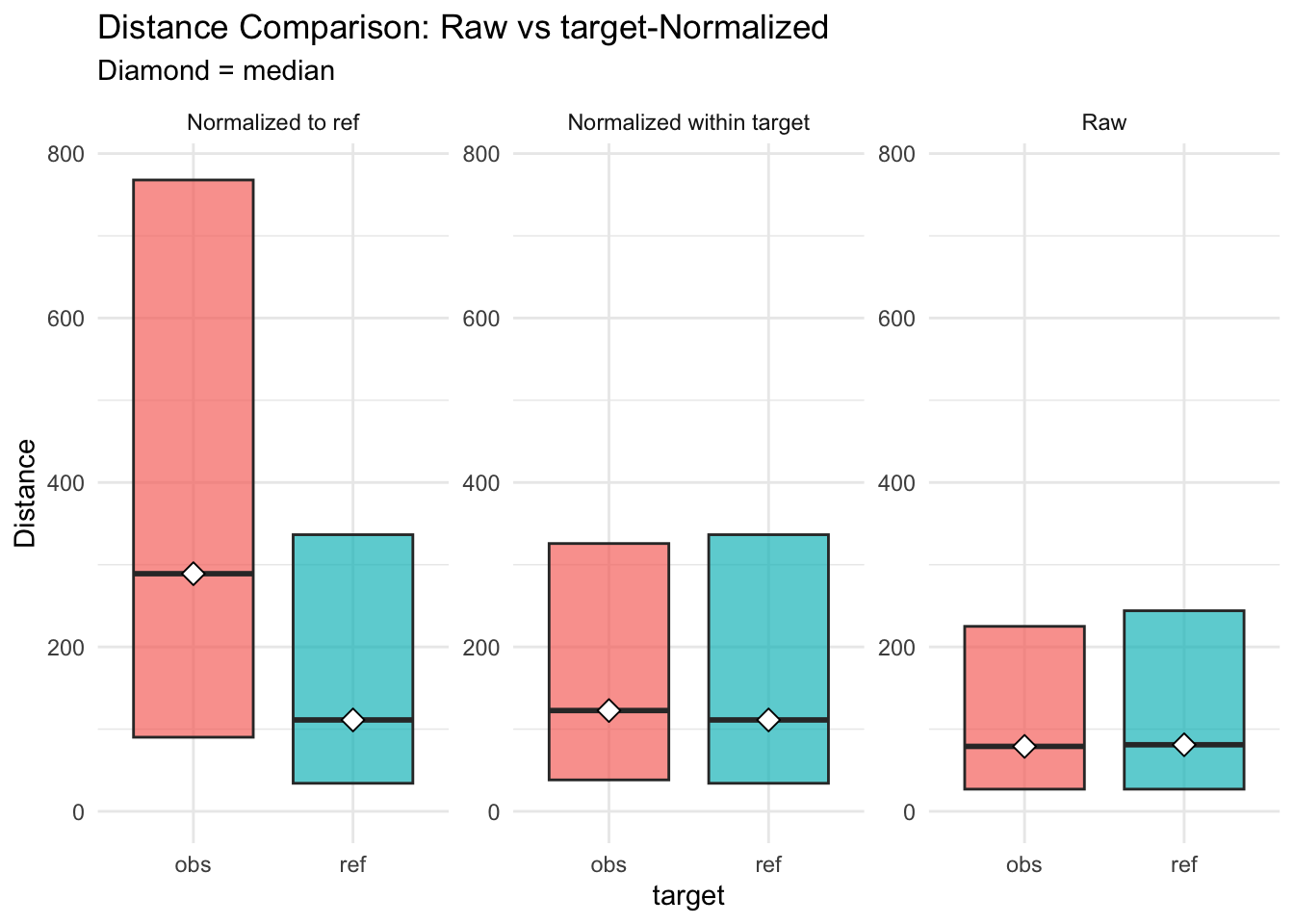
Figure 3.20: distances normalised vs. raw
3.6 evaluation model: 6
3.6.2 parameter setting
## value
## norm_target
## det.t TRUE
## limit TRUE
## author TRUE
## url TRUE
## embed1 TRUE
## embed2 f
## range1 TRUE
## range2 f
## rel FALSE
## lme FALSE
## lemma FALSE3.6.3 anova analysis
3.6.3.1 anova plain
formula: [dist ~ target*q*det]
## Df Sum Sq Mean Sq F value Pr(>F)
## target 1 3284330 3284330 84.1223 < 2.2e-16 ***
## q 5 1633205 326641 8.3663 6.39e-08 ***
## det 1 431404 431404 11.0496 0.0008873 ***
## target:q 5 441118 88224 2.2597 0.0457798 *
## target:det 1 16732 16732 0.4286 0.5126999
## q:det 2 25549 12774 0.3272 0.7209470
## target:q:det 1 6009 6009 0.1539 0.6948226
## Residuals 126209 4927490433 39042
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.6.3.2 anova of linear regression model
[anova(summary(lmer))]
## Type III Analysis of Variance Table with Satterthwaite's method
## Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
## target 218 218 1 17034 0.0061 0.9377
## q 109358 21872 5 124317 0.6129 0.6901
## det 20678 20678 1 121247 0.5794 0.4465
## range 15332432 15332432 1 912 429.6377 <2e-16 ***
## embed.score 77286239 77286239 1 105351 2165.6761 <2e-16 ***
## target:q 304923 60985 5 125126 1.7089 0.1287
## target:det 17833 17833 1 124982 0.4997 0.4796
## q:det 37151 18576 2 123066 0.5205 0.5942
## target:q:det 23985 23985 1 124972 0.6721 0.4123
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 13.6.3.3 linear regression coefficients
formula: [dist ~ target*q*det+(1|aut_id)+range+(embed.score)+(1|url_id)]
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: eval(expr(lmeform))
## Data: dfa
##
## REML criterion at convergence: 1685333
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.0402 -0.6622 -0.3317 0.3419 4.1697
##
## Random effects:
## Groups Name Variance Std.Dev.
## aut_id (Intercept) 1394 37.34
## url_id (Intercept) 1072 32.74
## Residual 35687 188.91
## Number of obs: 126226, groups: aut_id, 8238; url_id, 2145
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.533e+02 3.618e+00 1.966e+04 70.000 < 2e-16 ***
## targetref 1.326e+00 2.954e+00 1.890e+03 0.449 0.65362
## qb -8.195e+00 1.300e+01 1.239e+05 -0.630 0.52845
## qc -8.144e+00 4.675e+00 1.243e+05 -1.742 0.08150 .
## qd -1.117e+02 1.902e+02 1.212e+05 -0.587 0.55726
## qe 1.392e+01 3.248e+00 1.256e+05 4.285 1.83e-05 ***
## qf -6.628e+00 4.145e+00 1.253e+05 -1.599 0.10981
## det 3.793e+00 4.058e+00 1.245e+05 0.935 0.35005
## range 1.535e-02 7.406e-04 9.124e+02 20.728 < 2e-16 ***
## embed.score -3.110e+02 6.682e+00 1.054e+05 -46.537 < 2e-16 ***
## targetref:qb 4.017e+00 1.464e+01 1.244e+05 0.274 0.78373
## targetref:qc 4.577e+00 1.089e+01 1.253e+05 0.420 0.67442
## targetref:qd -2.061e+00 1.069e+01 1.253e+05 -0.193 0.84707
## targetref:qe -2.134e+01 8.099e+00 1.255e+05 -2.635 0.00841 **
## targetref:qf 8.889e+00 1.031e+01 1.254e+05 0.862 0.38849
## targetref:det 1.178e+00 9.236e+00 1.253e+05 0.127 0.89855
## qb:det 1.714e+01 1.428e+01 1.239e+05 1.200 0.23002
## qd:det 1.126e+02 1.902e+02 1.212e+05 0.592 0.55380
## targetref:qb:det -1.710e+01 2.086e+01 1.250e+05 -0.820 0.41233
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## fit warnings:
## fixed-effect model matrix is rank deficient so dropping 7 columns / coefficients
## Some predictor variables are on very different scales: consider rescaling3.6.4 plots
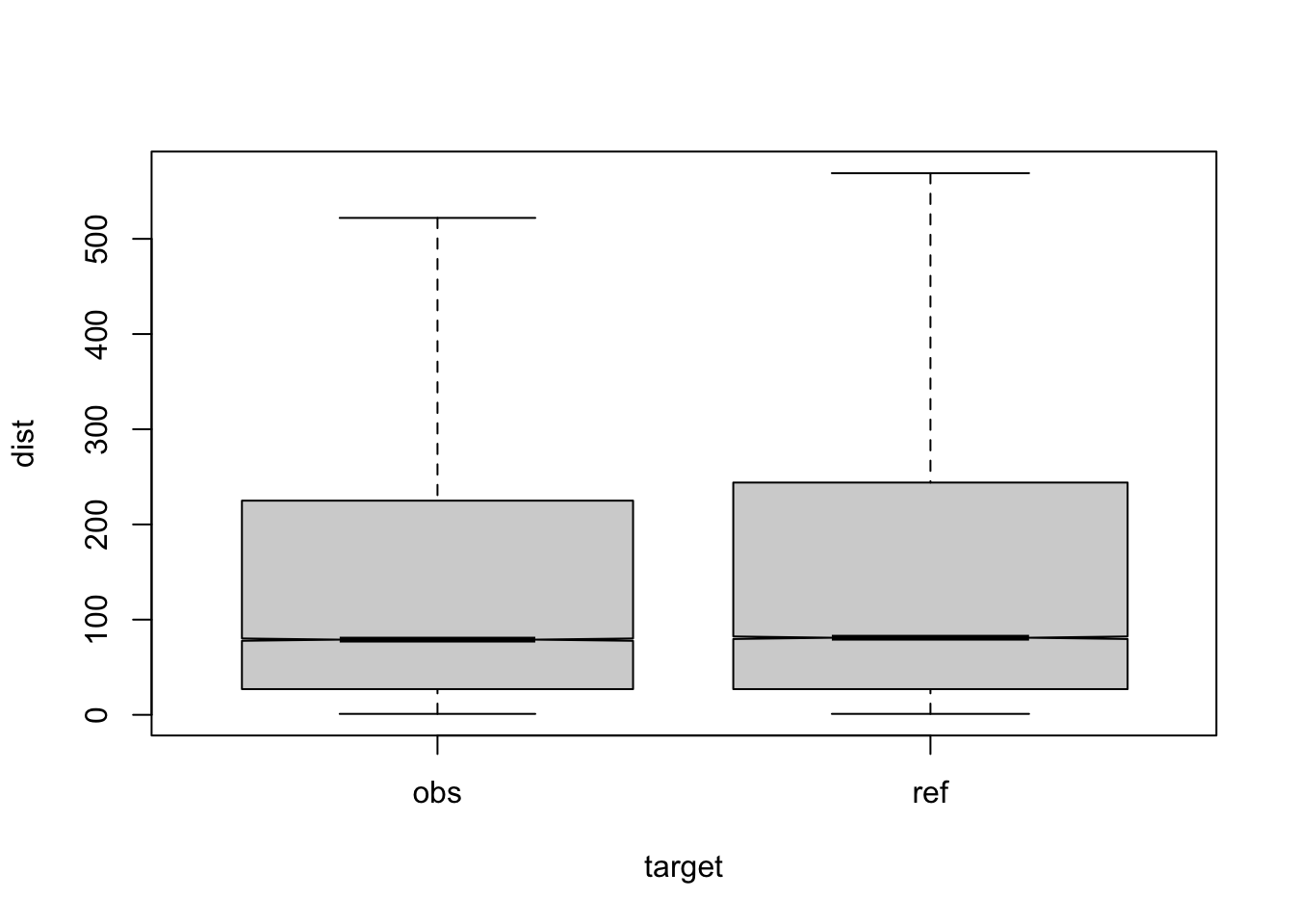
Figure 3.21: compare distances by corpus, not normalised, distance ceiling =outliers removed
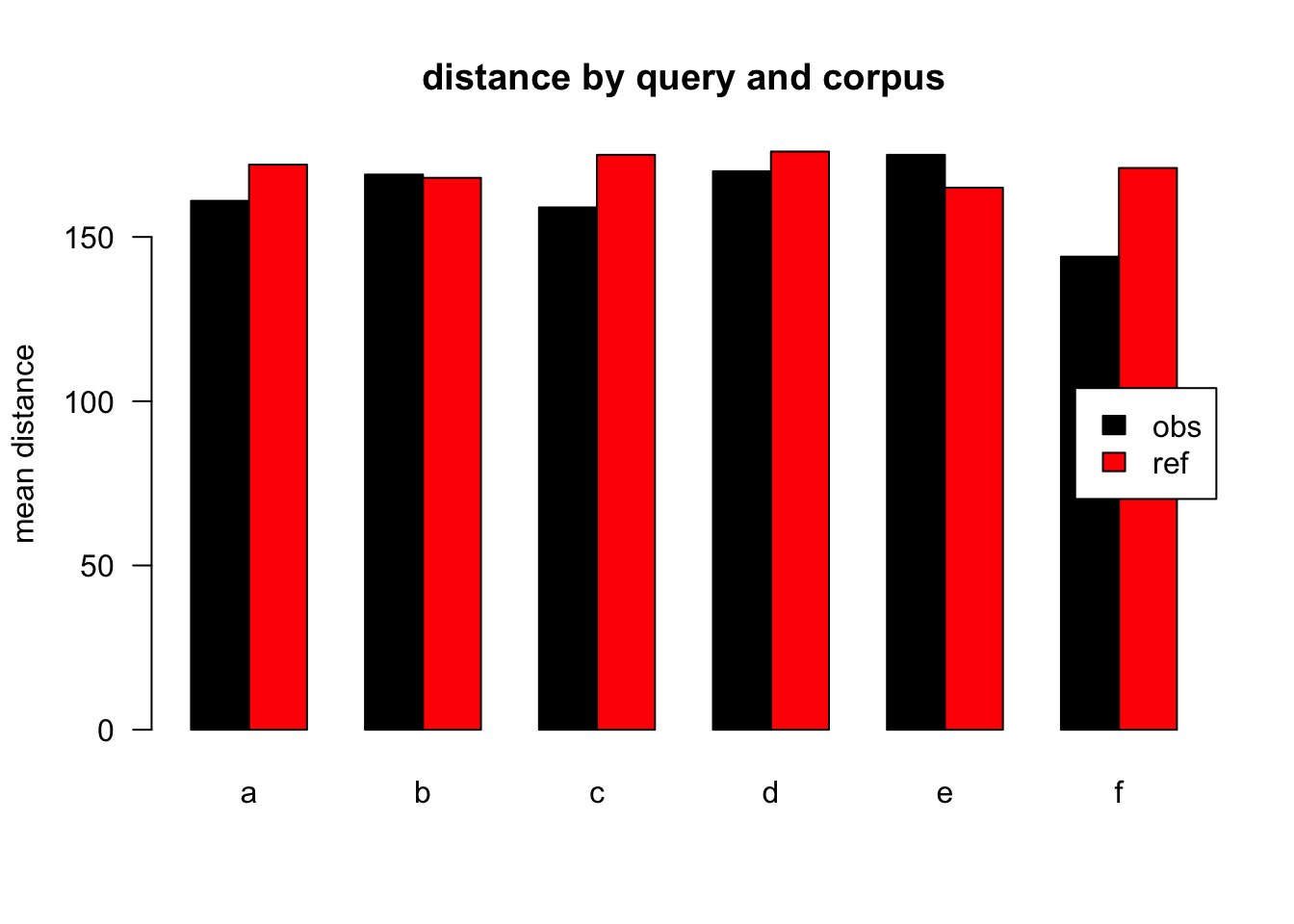
Figure 3.22: mean distances over query/corpus, not normalised, distance ceiling =outliers removed
| target | q | n | mean | median |
|---|---|---|---|---|
| obs | a | 42836 | 161 | 77 |
| ref | a | 58615 | 172 | 81 |
| obs | b | 2116 | 169 | 109 |
| ref | b | 1130 | 168 | 78 |
| obs | c | 5770 | 159 | 75 |
| ref | c | 1274 | 175 | 84 |
| obs | d | 5654 | 170 | 86 |
| ref | d | 1525 | 176 | 83 |
| obs | e | 3911 | 175 | 92 |
| ref | e | 671 | 165 | 71 |
| obs | f | 2311 | 144 | 62 |
| ref | f | 413 | 171 | 82 |
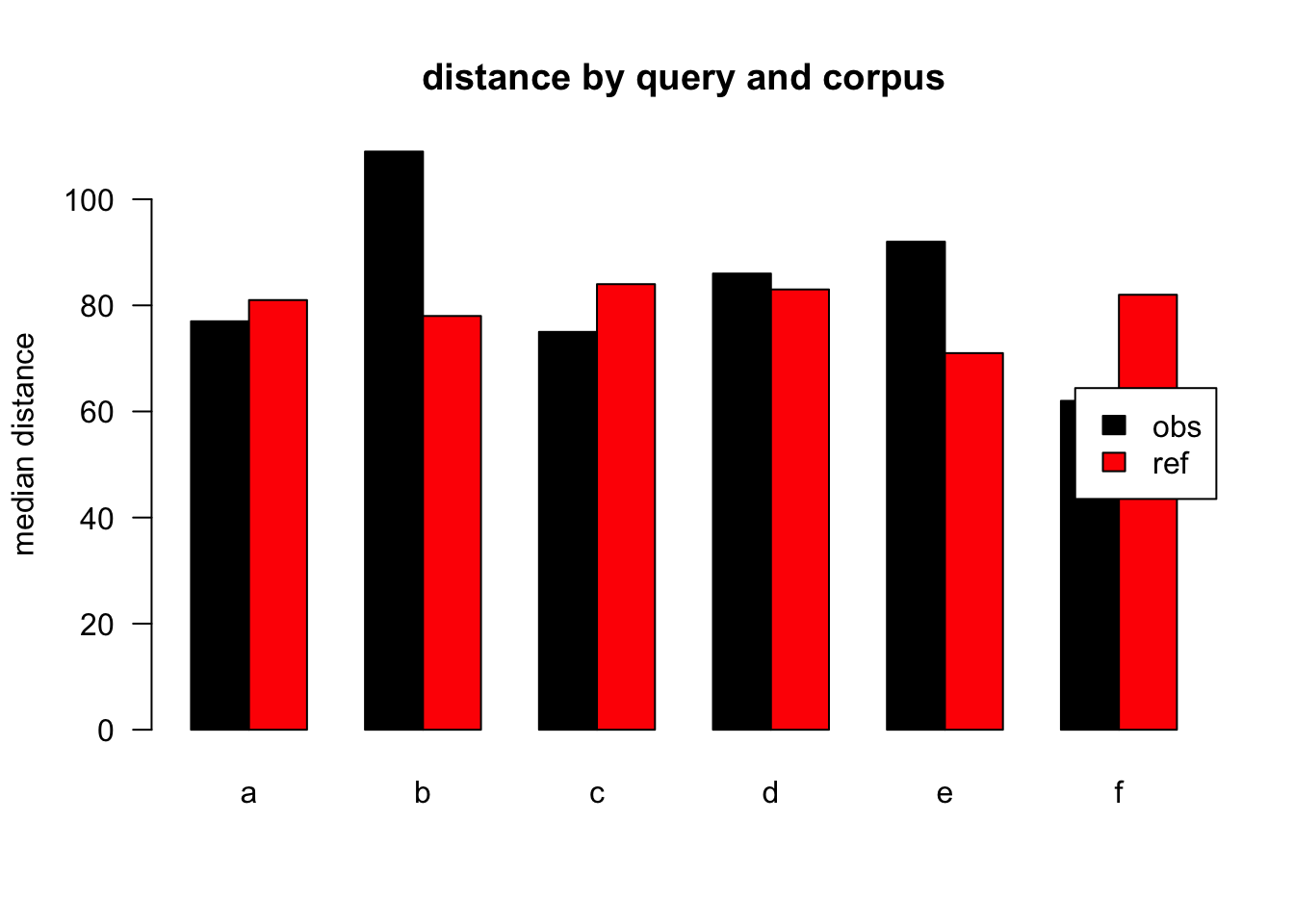
Figure 3.23: median distances over query/corpus, not normalised, distance ceiling =outliers removed
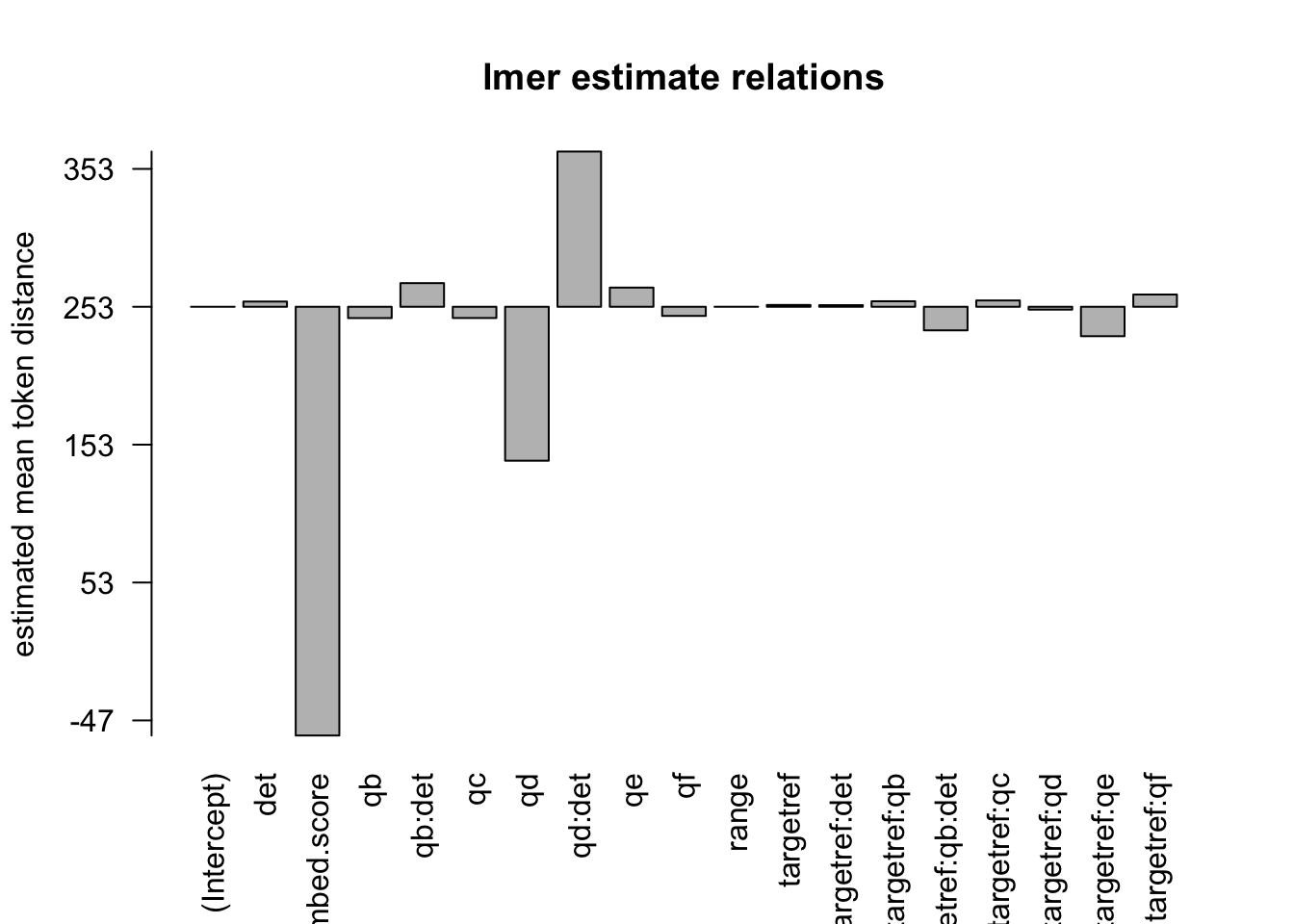
Figure 3.24: distances relation, not normalised, distance ceiling =outliers removed
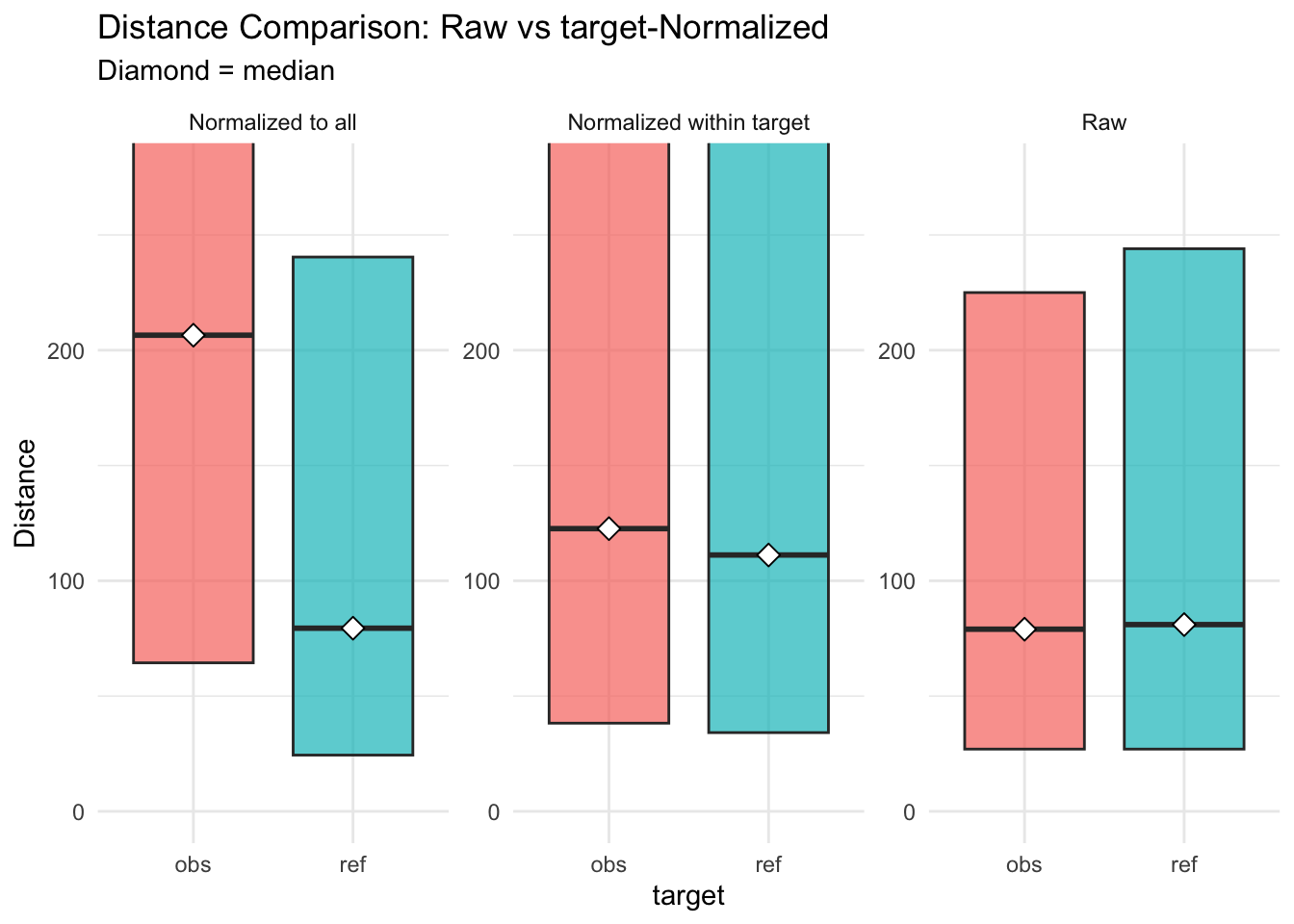
Figure 3.25: distances normalised vs. raw
3.7 Selbständigkeit: benutzte Hilfestellung
In der vorliegenden Arbeit wurden keinerlei nicht erlaubte Hilfsmittel zur Erstellung von Inhalten verwendet. Die Benutzung von KI beschränkt sich auf (Tabelle):
| Hilfsmittel | Verwendung |
|---|---|
| github copilot | Hilfe bei der Skripterstellung (R, Python) zur Programmierung der Distanzenberechnung, semantic embeddings und statistischen Auswertung |
| chatgpt.com | dito |
| claude.ai | dito |
| deepseek.com | dito |
| nomic-embed-text (model) | calculate semantic embeddings |
3.8 references
literature used and alii…